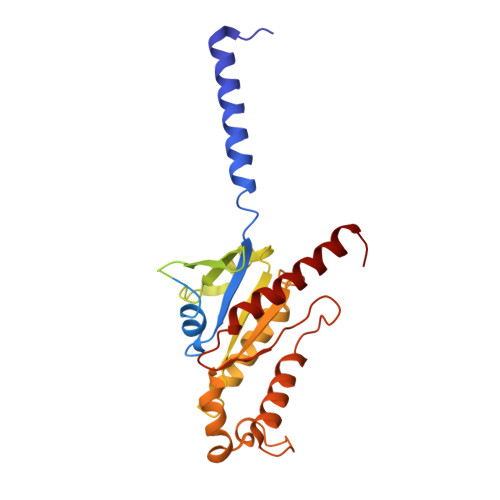
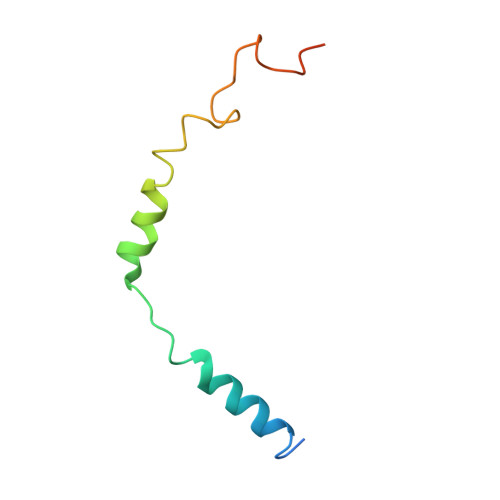
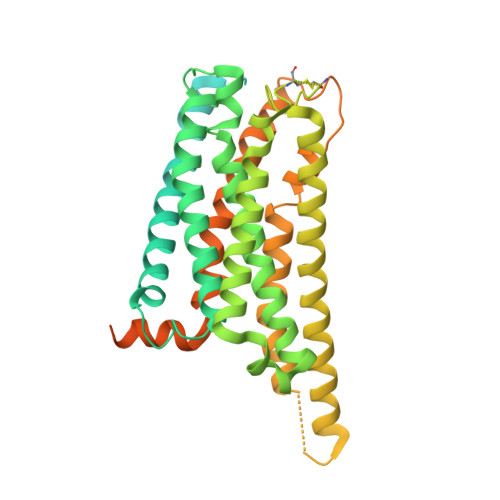
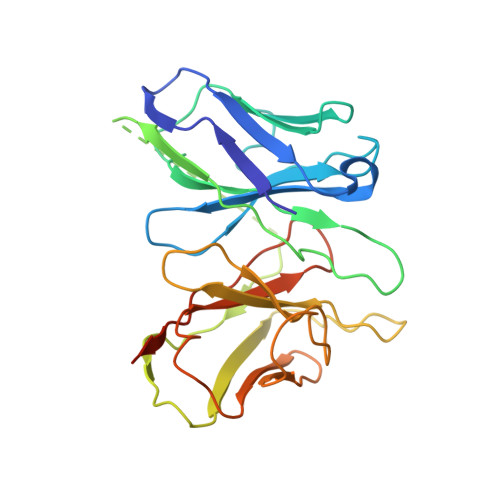
Structure of a Signaling Cannabinoid Receptor 1-G Protein Complex.
Krishna Kumar, K., Shalev-Benami, M., Robertson, M.J., Hu, H., Banister, S.D., Hollingsworth, S.A., Latorraca, N.R., Kato, H.E., Hilger, D., Maeda, S., Weis, W.I., Farrens, D.L., Dror, R.O., Malhotra, S.V., Kobilka, B.K., Skiniotis, G.(2019) Cell 176: 448
- PubMed: 30639101
- DOI: https://doi.org/10.1016/j.cell.2018.11.040
- Primary Citation of Related Structures:
6N4B - PubMed Abstract:
Cannabis elicits its mood-enhancing and analgesic effects through the cannabinoid receptor 1 (CB1), a G protein-coupled receptor (GPCR) that signals primarily through the adenylyl cyclase-inhibiting heterotrimeric G protein G i . Activation of CB1-G i signaling pathways holds potential for treating a number of neurological disorders and is thus crucial to understand the mechanism of G i activation by CB1. Here, we present the structure of the CB1-G i signaling complex bound to the highly potent agonist MDMB-Fubinaca (FUB), a recently emerged illicit synthetic cannabinoid infused in street drugs that have been associated with numerous overdoses and fatalities. The structure illustrates how FUB stabilizes the receptor in an active state to facilitate nucleotide exchange in G i . The results compose the structural framework to explain CB1 activation by different classes of ligands and provide insights into the G protein coupling and selectivity mechanisms adopted by the receptor.
- Department of Molecular and Cellular Physiology, Stanford University School of Medicine, 279 Campus Drive, Stanford, CA 94305, USA.
Organizational Affiliation: