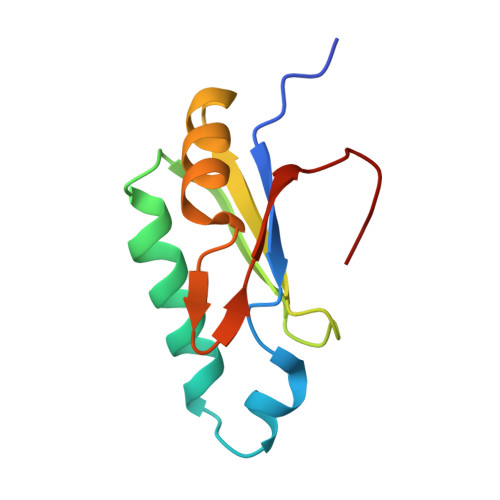
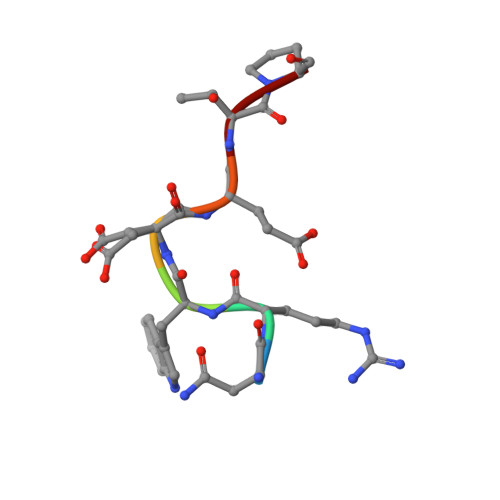
The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
Loerch, S., Leach, J.R., Horner, S.W., Maji, D., Jenkins, J.L., Pulvino, M.J., Kielkopf, C.L.(2019) J Biological Chem 294: 2892-2902
- PubMed: 30567737
- DOI: https://doi.org/10.1074/jbc.RA118.006764
- Primary Citation of Related Structures:
6N3D, 6N3E, 6N3F - PubMed Abstract:
The transcription elongation and pre-mRNA splicing factor Tat-SF1 associates with the U2 small nuclear ribonucleoprotein (snRNP) of the spliceosome. However, the direct binding partner and underlying interactions mediating the Tat-SF1-U2 snRNP association remain unknown. Here, we identified SF3b1 as a Tat-SF1-interacting subunit of the U2 snRNP. Our 1.1 Å resolution crystal structure revealed that Tat-SF1 contains a U2AF homology motif (UHM) protein-protein interaction module. We demonstrated that Tat-SF1 preferentially and directly binds the SF3b1 subunit compared with other U2AF ligand motif (ULM)-containing splicing factors, and further established that SF3b1 association depends on the integrity of the Tat-SF1 UHM. We next compared the Tat-SF1-binding affinities for each of the five known SF3b1 ULMs and then determined the structures of representative high- and low-affinity SF3b1 ULM complexes with the Tat-SF1 UHM at 1.9 Å and 2.1 Å resolutions, respectively. These structures revealed a canonical UHM-ULM interface, comprising a Tat-SF1 binding pocket for a ULM tryptophan (SF3b1 Trp 338 ) and electrostatic interactions with a basic ULM tail. Importantly, we found that SF3b1 regulates Tat-SF1 levels and that these two factors influence expression of overlapping representative transcripts, consistent with a functional partnership of Tat-SF1 and SF3b1. Altogether, these results define a new molecular interface of the Tat-SF1-U2 snRNP complex for gene regulation.
- From the Center for RNA Biology, Department of Biochemistry and Biophysics, University of Rochester School of Medicine and Dentistry, Rochester, New York 14642.
Organizational Affiliation: