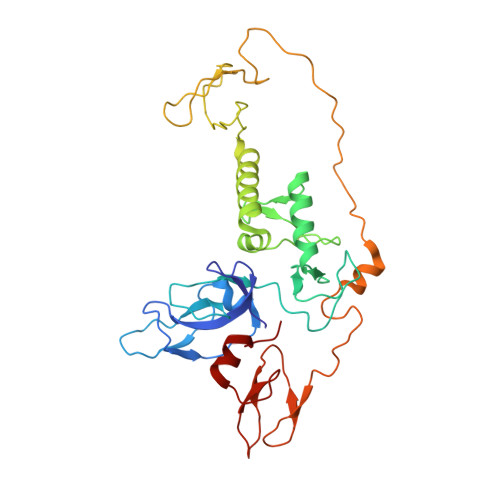
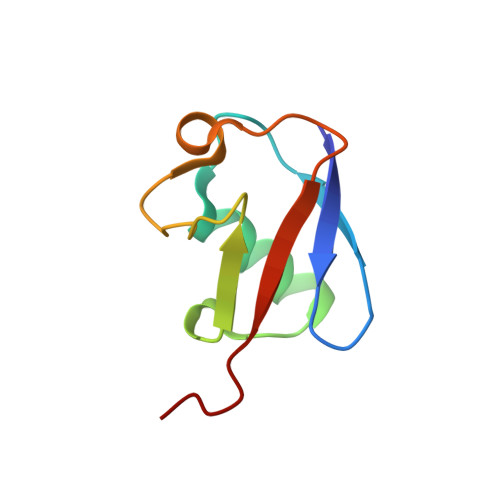
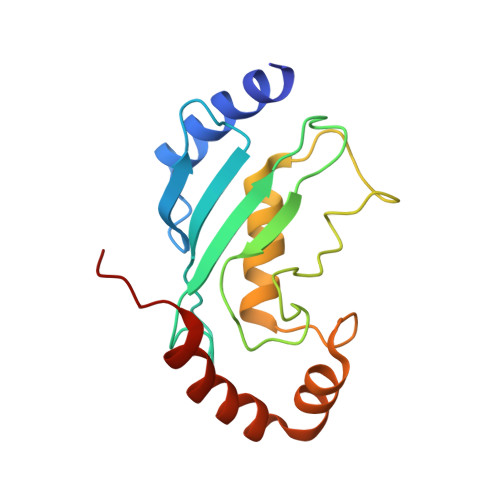
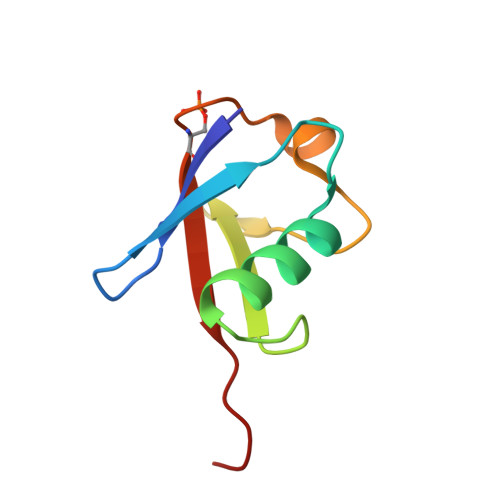
Synergistic recruitment of UbcH7~Ub and phosphorylated Ubl domain triggers parkin activation.
Condos, T.E., Dunkerley, K.M., Freeman, E.A., Barber, K.R., Aguirre, J.D., Chaugule, V.K., Xiao, Y., Konermann, L., Walden, H., Shaw, G.S.(2018) EMBO J 37
- PubMed: 30446597
- DOI: https://doi.org/10.15252/embj.2018100014
- Primary Citation of Related Structures:
6N13 - PubMed Abstract:
The E3 ligase parkin ubiquitinates outer mitochondrial membrane proteins during oxidative stress and is linked to early-onset Parkinson's disease. Parkin is autoinhibited but is activated by the kinase PINK1 that phosphorylates ubiquitin leading to parkin recruitment, and stimulates phosphorylation of parkin's N-terminal ubiquitin-like (pUbl) domain. How these events alter the structure of parkin to allow recruitment of an E2~Ub conjugate and enhanced ubiquitination is an unresolved question. We present a model of an E2~Ub conjugate bound to the phospho-ubiquitin-loaded C-terminus of parkin, derived from NMR chemical shift perturbation experiments. We show the UbcH7~Ub conjugate binds in the open state whereby conjugated ubiquitin binds to the RING1/IBR interface. Further, NMR and mass spectrometry experiments indicate the RING0/RING2 interface is re-modelled, remote from the E2 binding site, and this alters the reactivity of the RING2(Rcat) catalytic cysteine, needed for ubiquitin transfer. Our experiments provide evidence that parkin phosphorylation and E2~Ub recruitment act synergistically to enhance a weak interaction of the pUbl domain with the RING0 domain and rearrange the location of the RING2(Rcat) domain to drive parkin activity.
- Department of Biochemistry, The University of Western Ontario, London, ON, Canada.
Organizational Affiliation: