Structure of Tdp1 catalytic domain in complex with compound XZ520
Lountos, G.T., Zhao, X.Z., Kiselev, E., Tropea, J.E., Needle, D., Burke Jr., T.R., Pommier, Y., Waugh, D.S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tyrosyl-DNA phosphodiesterase 1 | 461 | Homo sapiens | Mutation(s): 0 Gene Names: TDP1 EC: 3.1.4 | 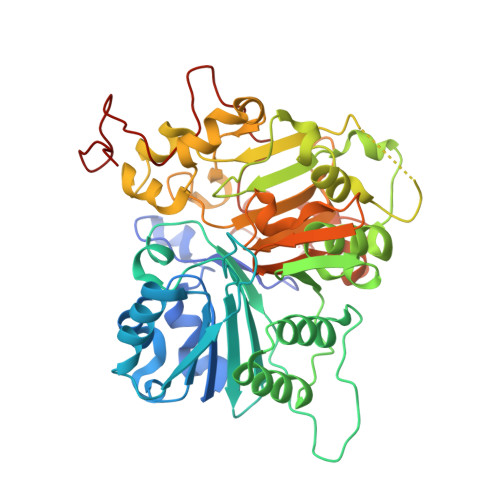 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9NUW8 (Homo sapiens) Explore Q9NUW8 Go to UniProtKB: Q9NUW8 | |||||
PHAROS: Q9NUW8 GTEx: ENSG00000042088 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9NUW8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| K7D Query on K7D | C [auth A] | 4-oxo-8-phenyl-1,4-dihydroquinoline-3-carboxylic acid C16 H11 N O3 IYZQCKZKSCOMCY-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | D [auth A], E [auth A], F [auth A], G [auth B], H [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 50.065 | α = 90 |
| b = 105.586 | β = 90 |
| c = 194.67 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| PHASER | phasing |