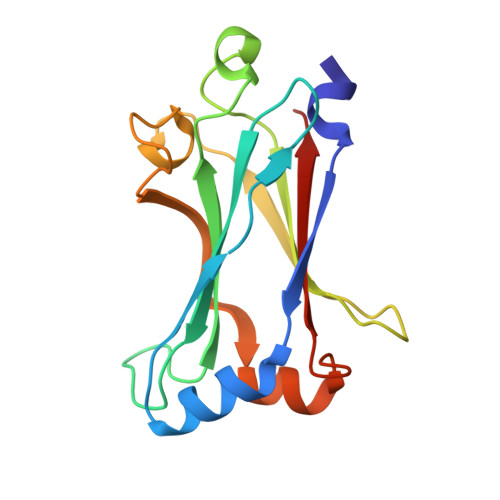
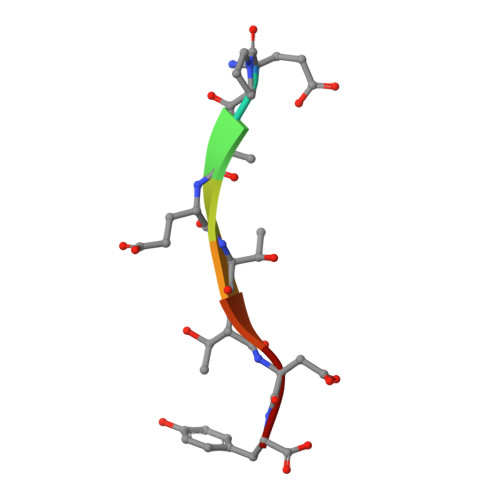
Modular Architecture of the STING C-Terminal Tail Allows Interferon and NF-kappa B Signaling Adaptation.
de Oliveira Mann, C.C., Orzalli, M.H., King, D.S., Kagan, J.C., Lee, A.S.Y., Kranzusch, P.J.(2019) Cell Rep 27: 1165-1175.e5
- PubMed: 31018131
- DOI: https://doi.org/10.1016/j.celrep.2019.03.098
- Primary Citation of Related Structures:
6MYD - PubMed Abstract:
Stimulator of interferon genes (STING) is a key regulator of type I interferon and pro-inflammatory responses during infection, cellular stress, and cancer. Here, we reveal a mechanism for how STING balances activation of IRF3- and NF-κB-dependent transcription and discover that acquisition of discrete signaling modules in the vertebrate STING C-terminal tail (CTT) shapes downstream immunity. As a defining example, we identify a motif appended to the CTT of zebrafish STING that inverts the typical vertebrate signaling response and results in dramatic NF-κB activation and weak IRF3-interferon signaling. We determine a co-crystal structure that explains how this CTT sequence recruits TRAF6 as a new binding partner and demonstrate that the minimal motif is sufficient to reprogram human STING and immune activation in macrophage cells. Together, our results define the STING CTT as a linear signaling hub that can acquire modular motifs to readily adapt downstream immunity.
- Department of Microbiology, Harvard Medical School, Boston, MA 02115, USA; Department of Cancer Immunology and Virology, Dana-Farber Cancer Institute, Boston, MA 02115, USA.
Organizational Affiliation: