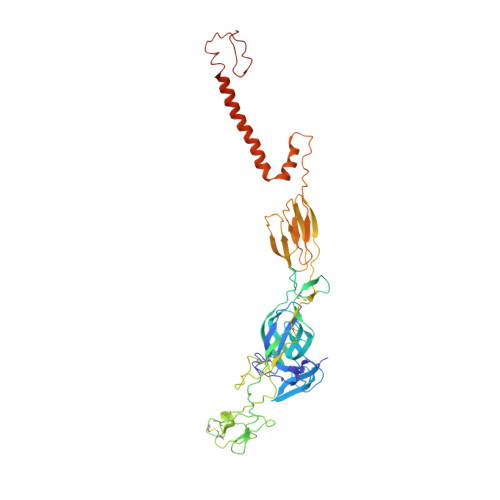
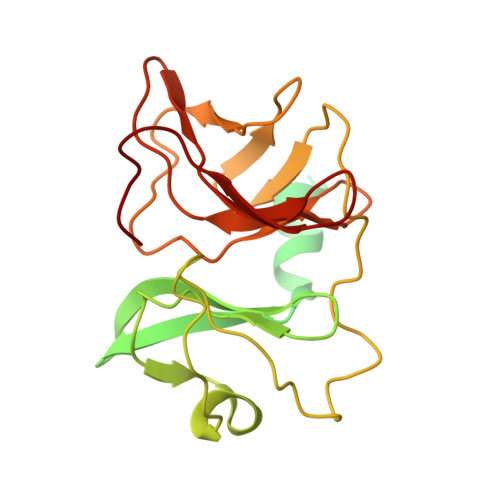
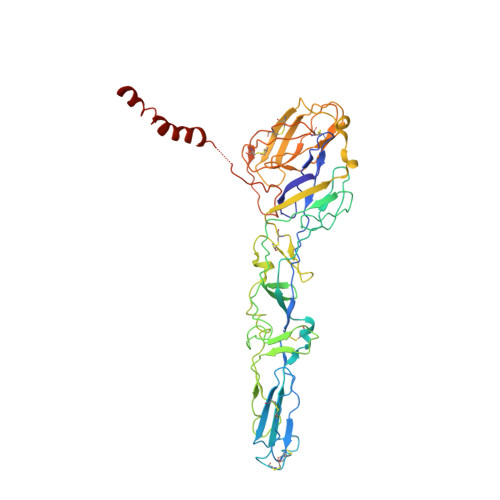Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Hasan, S.S., Sun, C., Kim, A.S., Watanabe, Y., Chen, C.L., Klose, T., Buda, G., Crispin, M., Diamond, M.S., Klimstra, W.B., Rossmann, M.G.(2018) Cell Rep 25: 3136-3147.e5
- PubMed: 30540945
- DOI: https://doi.org/10.1016/j.celrep.2018.11.067
- Primary Citation of Related Structures:
6MUI, 6MW9, 6MWC, 6MWV, 6MWX, 6MX4, 6MX7 - PubMed Abstract:
Alphaviruses are enveloped pathogens that cause arthritis and encephalitis. Here, we report a 4.4-Å cryoelectron microscopy (cryo-EM) structure of eastern equine encephalitis virus (EEEV), an alphavirus that causes fatal encephalitis in humans. Our analysis provides insights into viral entry into host cells. The envelope protein E2 showed a binding site for the cellular attachment factor heparan sulfate. The presence of a cryptic E2 glycan suggests how EEEV escapes surveillance by lectin-expressing myeloid lineage cells, which are sentinels of the immune system. A mechanism for nucleocapsid core release and disassembly upon viral entry was inferred based on pH changes and capsid dissociation from envelope proteins. The EEEV capsid structure showed a viral RNA genome binding site adjacent to a ribosome binding site for viral genome translation following genome release. Using five Fab-EEEV complexes derived from neutralizing antibodies, our investigation provides insights into EEEV host cell interactions and protective epitopes relevant to vaccine design.
- Department of Biological Sciences, Purdue University, West Lafayette, IN 47907, USA.
Organizational Affiliation: