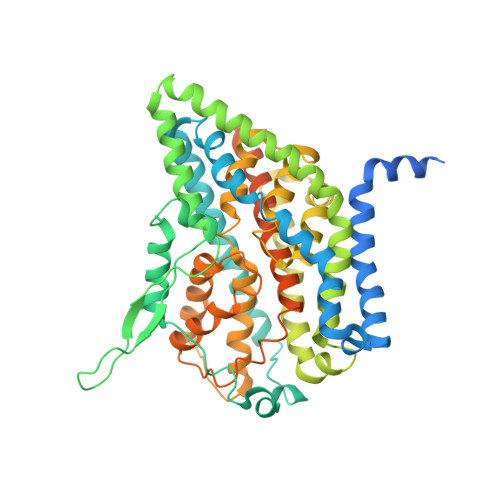
Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Yu, X., Plotnikova, O., Bonin, P.D., Subashi, T.A., McLellan, T.J., Dumlao, D., Che, Y., Dong, Y.Y., Carpenter, E.P., West, G.M., Qiu, X., Culp, J.S., Han, S.(2019) Elife 8
- PubMed: 31580259
- DOI: https://doi.org/10.7554/eLife.48120
- Primary Citation of Related Structures:
6MP6, 6MPB - PubMed Abstract:
Alanine-serine-cysteine transporter 2 (ASCT2, SLC1A5) is the primary transporter of glutamine in cancer cells and regulates the mTORC1 signaling pathway. The SLC1A5 function involves finely tuned orchestration of two domain movements that include the substrate-binding transport domain and the scaffold domain. Here, we present cryo-EM structures of human SLC1A5 and its complex with the substrate, L-glutamine in an outward-facing conformation. These structures reveal insights into the conformation of the critical ECL2a loop which connects the two domains, thus allowing rigid body movement of the transport domain throughout the transport cycle. Furthermore, the structures provide new insights into substrate recognition, which involves conformational changes in the HP2 loop. A putative cholesterol binding site was observed near the domain interface in the outward-facing state. Comparison with the previously determined inward-facing structure of SCL1A5 provides a basis for a more integrated understanding of substrate recognition and transport mechanism in the SLC1 family.
- Medicine Design, Pfizer Inc, Groton, United States.
Organizational Affiliation: