cAMP-dependent protein kinase A in complex with RyR2 peptide (2799-2810)
Haji-Ghassemi, O., Yuchi, Z., van Petegem, F.(2019) Mol Cell
Experimental Data Snapshot
Starting Model: experimental
View more details
(2019) Mol Cell
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| cAMP-dependent protein kinase catalytic subunit alpha | A, C [auth D] | 339 | Mus musculus | Mutation(s): 0 Gene Names: Prkaca, Pkaca EC: 2.7.11.11 | 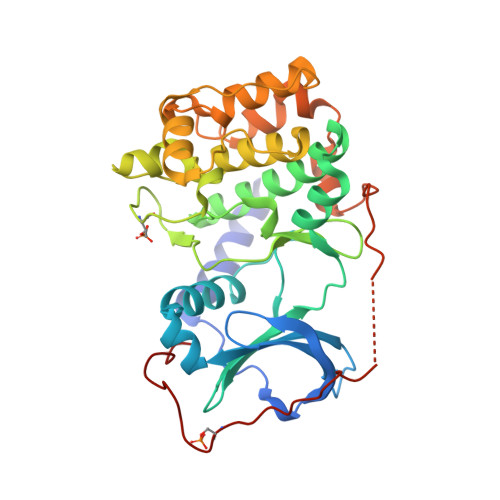 |
UniProt | |||||
Find proteins for P05132 (Mus musculus) Explore P05132 Go to UniProtKB: P05132 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P05132 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ryanodine receptor 2 | B [auth C], D [auth E] | 209 | Mus musculus | Mutation(s): 2 Gene Names: Ryr2 | 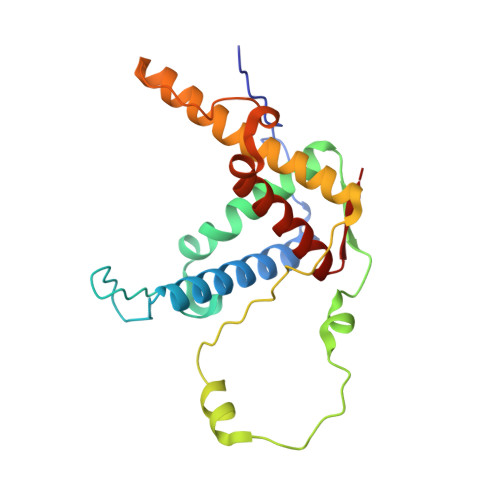 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for E9Q401 (Mus musculus) Explore E9Q401 Go to UniProtKB: E9Q401 | |||||
IMPC: MGI:99685 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | E9Q401 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ANP (Subject of Investigation/LOI) Query on ANP | J [auth A], Q [auth D] | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER C10 H17 N6 O12 P3 PVKSNHVPLWYQGJ-KQYNXXCUSA-N |  | ||
| EDO Query on EDO | K [auth A] L [auth C] S [auth D] T [auth D] U [auth D] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| ACY Query on ACY | G [auth A] | ACETIC ACID C2 H4 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-N |  | ||
| FMT Query on FMT | E [auth A] F [auth A] H [auth A] I [auth A] M [auth D] | FORMIC ACID C H2 O2 BDAGIHXWWSANSR-UHFFFAOYSA-N |  | ||
| CL Query on CL | R [auth D], W [auth E] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Modified Residues 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| SEP Query on SEP | A, C [auth D] | L-PEPTIDE LINKING | C3 H8 N O6 P |  | SER |
| TPO Query on TPO | A, C [auth D] | L-PEPTIDE LINKING | C4 H10 N O6 P |  | THR |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 58.337 | α = 90 |
| b = 67.56 | β = 90.73 |
| c = 176.852 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data scaling |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Canadian Institutes of Health Research (CIHR) | Canada | PJT 153305 |