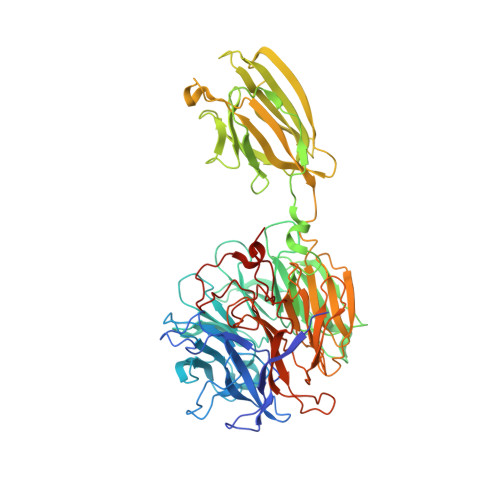
The 1.9 angstrom crystal structure of the extracellular matrix protein Bap1 fromVibrio choleraeprovides insights into bacterial biofilm adhesion.
Kaus, K., Biester, A., Chupp, E., Lu, J., Visudharomn, C., Olson, R.(2019) J Biological Chem 294: 14499-14511
- PubMed: 31439670
- DOI: https://doi.org/10.1074/jbc.RA119.008335
- Primary Citation of Related Structures:
6MLT - PubMed Abstract:
Growth of the cholera bacterium Vibrio cholerae in a biofilm community contributes to both its pathogenicity and survival in aquatic environmental niches. The major components of V. cholerae biofilms include V ibrio p oly s accharide (VPS) and the extracellular matrix proteins RbmA, RbmC, and Bap1. To further elucidate the previously observed overlapping roles of Bap1 and RbmC in biofilm architecture and surface attachment, here we investigated the structural and functional properties of Bap1. Soluble expression of Bap1 was possible only after the removal of an internal 57-amino-acid-long hydrophobic insertion sequence. The crystal structure of Bap1 at 1.9 Å resolution revealed a two-domain assembly made up of an eight-bladed β-propeller interrupted by a β-prism domain. The structure also revealed metal-binding sites within canonical calcium blade motifs, which appear to have structural rather than functional roles. Contrary to results previously observed with RbmC, the Bap1 β-prism domain did not exhibit affinity for complex N -glycans, suggesting an altered role of this domain in biofilm-surface adhesion. Native polyacrylamide gel shift analysis did suggest that Bap1 exhibits lectin activity with a preference for anionic or linear polysaccharides. Our results suggest a model for V. cholerae biofilms in which Bap1 and RbmC play dominant but differing adhesive roles in biofilms, allowing bacterial attachment to diverse environmental or host surfaces.
- Department of Molecular Biology and Biochemistry, Molecular Biophysics Program, Wesleyan University, Middletown, Connecticut 06459.
Organizational Affiliation: