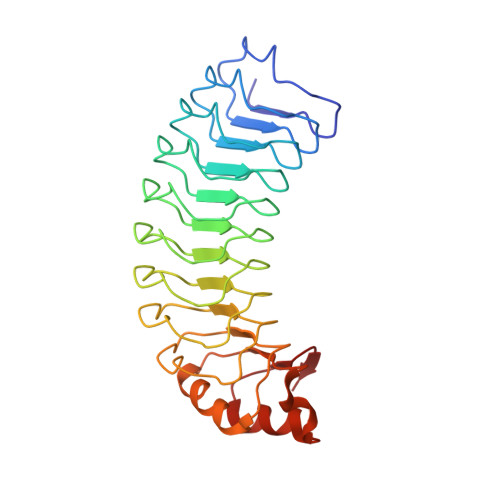
The structure of SDS22 provides insights into the mechanism of heterodimer formation with PP1.
Choy, M.S., Bolik-Coulon, N., Archuleta, T.L., Peti, W., Page, R.(2018) Acta Crystallogr F Struct Biol Commun 74: 817-824
- PubMed: 30511677
- DOI: https://doi.org/10.1107/S2053230X18016503
- Primary Citation of Related Structures:
6MKY - PubMed Abstract:
Protein phosphatase 1 (PP1) dephosphorylates hundreds of key biological targets by associating with nearly 200 regulatory proteins to form highly specific holoenzymes. The vast majority of regulators are intrinsically disordered proteins (IDPs) and bind PP1 via short linear motifs within their intrinsically disordered regions. One of the most ancient PP1 regulators is SDS22, a protein that is conserved from yeast to mammals. Sequence analysis of SDS22 revealed that it is a leucine-rich repeat (LRR) protein, suggesting that SDS22, unlike nearly every other known PP1 regulator, is not an IDP but instead is fully structured. Here, the 2.9 Å resolution crystal structure of human SDS22 in space group P2 1 2 1 2 1 is reported. SDS22 adopts an LRR fold with the horseshoe-like curvature typical for this family of proteins. The structure results in surfaces with distinct chemical characteristics that are likely to be critical for PP1 binding.
- Chemistry and Biochemistry, University of Arizona, 1041 East Lowell Drive, Biosciences West, Tucson, AZ 85281, USA.
Organizational Affiliation: