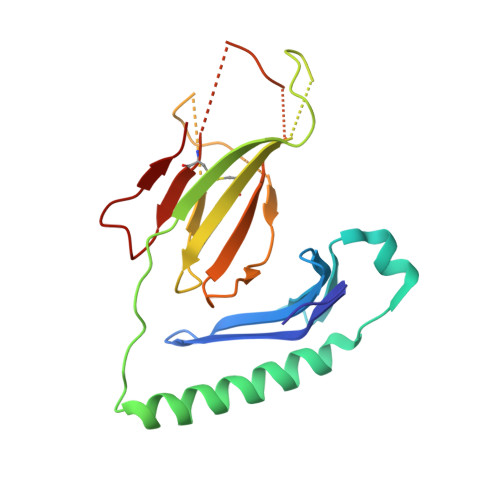
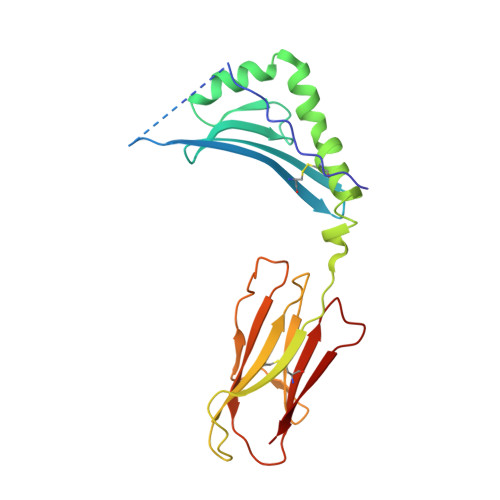
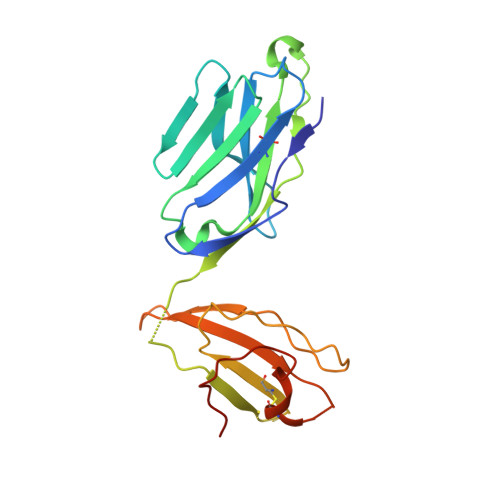
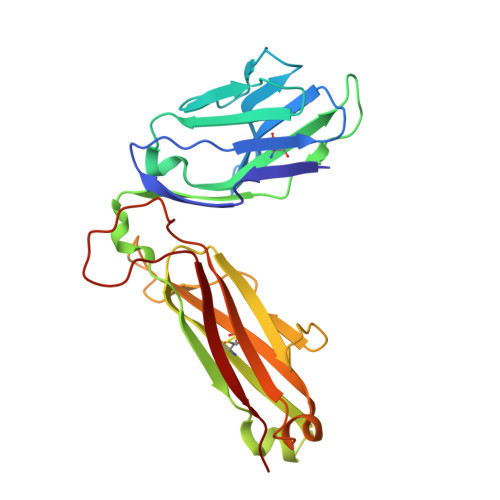
A temporal thymic selection switch and ligand binding kinetics constrain neonatal Foxp3+Tregcell development.
Stadinski, B.D., Blevins, S.J., Spidale, N.A., Duke, B.R., Huseby, P.G., Stern, L.J., Huseby, E.S.(2019) Nat Immunol 20: 1046-1058
- PubMed: 31209405
- DOI: https://doi.org/10.1038/s41590-019-0414-1
- Primary Citation of Related Structures:
6MKD, 6MKR, 6MNG, 6MNM, 6MNN, 6MNO - PubMed Abstract:
The neonatal thymus generates Foxp3 + regulatory T (tT reg ) cells that are critical in controlling immune homeostasis and preventing multiorgan autoimmunity. The role of antigen specificity on neonatal tT reg cell selection is unresolved. Here we identify 17 self-peptides recognized by neonatal tT reg cells, and reveal ligand specificity patterns that include self-antigens presented in an age- and inflammation-dependent manner. Fate-mapping studies of neonatal peptidyl arginine deiminase type IV (Padi4)-specific thymocytes reveal disparate fate choices. Neonatal thymocytes expressing T cell receptors that engage IA b -Padi4 with moderate dwell times within a conventional docking orientation are exported as tT reg cells. In contrast, Padi4-specific T cell receptors with short dwell times are expressed on CD4 + T cells, while long dwell times induce negative selection. Temporally, Padi4-specific thymocytes are subject to a developmental stage-specific change in negative selection, which precludes tT reg cell development. Thus, a temporal switch in negative selection and ligand binding kinetics constrains the neonatal tT reg selection window.
- Department of Pathology, University of Massachusetts Medical School, Worcester, MA, USA.
Organizational Affiliation: