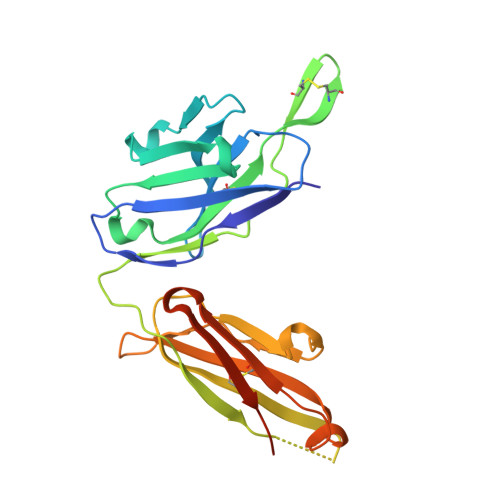
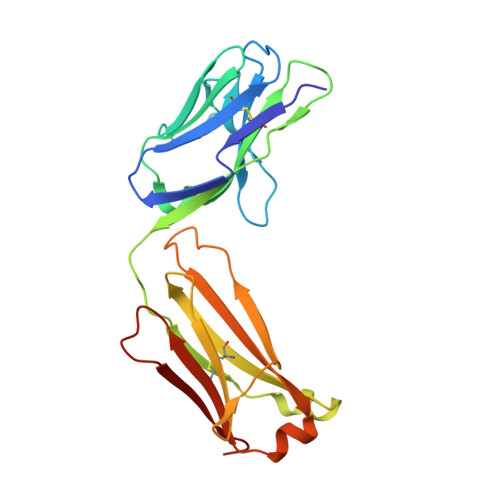
HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design.
Flyak, A.I., Ruiz, S., Colbert, M.D., Luong, T., Crowe Jr., J.E., Bailey, J.R., Bjorkman, P.J.(2018) Cell Host Microbe 24: 703-716.e3
- PubMed: 30439340
- DOI: https://doi.org/10.1016/j.chom.2018.10.009
- Primary Citation of Related Structures:
6MED, 6MEE, 6MEF, 6MEG, 6MEH, 6MEI, 6MEJ, 6MEK - PubMed Abstract:
Hepatitis C virus (HCV) vaccine efforts are hampered by the extensive genetic diversity of HCV envelope glycoproteins E1 and E2. Structures of broadly neutralizing antibodies (bNAbs) (e.g., HEPC3, HEPC74) isolated from individuals who spontaneously cleared HCV infection facilitate immunogen design to elicit antibodies against multiple HCV variants. However, challenges in expressing HCV glycoproteins previously limited bNAb-HCV structures to complexes with truncated E2 cores. Here we describe crystal structures of full-length E2 ectodomain complexes with HEPC3 and HEPC74, revealing lock-and-key antibody-antigen interactions, E2 regions (including a target of immunogen design) that were truncated or disordered in E2 cores, and an antibody CDRH3 disulfide motif that exhibits common interactions with a conserved epitope despite different bNAb-E2 binding orientations. The structures display unusual features relevant to common genetic signatures of HCV bNAbs and demonstrate extraordinary plasticity in antibody-antigen interactions. In addition, E2 variants that bind HEPC3/HEPC74-like germline precursors may represent candidate vaccine immunogens.
- Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, CA 91125, USA.
Organizational Affiliation: