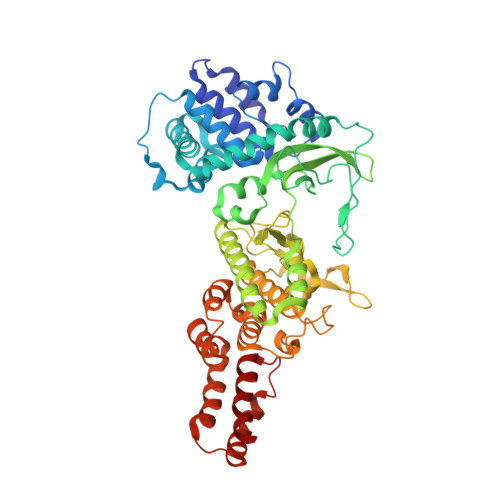
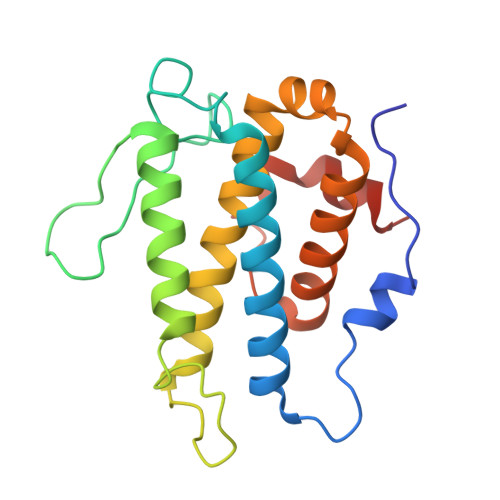
TheLegionellakinase LegK7 exploits the Hippo pathway scaffold protein MOB1A for allostery and substrate phosphorylation.
Lee, P.C., Beyrakhova, K., Xu, C., Boniecki, M.T., Lee, M.H., Onu, C.J., Grishin, A.M., Machner, M.P., Cygler, M.(2020) Proc Natl Acad Sci U S A 117: 14433-14443
- PubMed: 32513747
- DOI: https://doi.org/10.1073/pnas.2000497117
- Primary Citation of Related Structures:
6MCP, 6MCQ - PubMed Abstract:
During infection, the bacterial pathogen Legionella pneumophila manipulates a variety of host cell signaling pathways, including the Hippo pathway which controls cell proliferation and differentiation in eukaryotes. Our previous studies revealed that L. pneumophila encodes the effector kinase LegK7 which phosphorylates MOB1A, a highly conserved scaffold protein of the Hippo pathway. Here, we show that MOB1A, in addition to being a substrate of LegK7, also functions as an allosteric activator of its kinase activity. A crystallographic analysis of the LegK7-MOB1A complex revealed that the N-terminal half of LegK7 is structurally similar to eukaryotic protein kinases, and that MOB1A directly binds to the LegK7 kinase domain. Substitution of interface residues critical for complex formation abrogated allosteric activation of LegK7 both in vitro and within cells and diminished MOB1A phosphorylation. Importantly, the N-terminal extension (NTE) of MOB1A not only regulated complex formation with LegK7 but also served as a docking site for downstream substrates such as the transcriptional coregulator YAP1. Deletion of the NTE from MOB1A or addition of NTE peptides as binding competitors attenuated YAP1 recruitment to and phosphorylation by LegK7. By providing mechanistic insight into the formation and regulation of the LegK7-MOB1A complex, our study unravels a sophisticated molecular mimicry strategy that is used by L. pneumophila to take control of the host cell Hippo pathway.
- Division of Molecular and Cellular Biology, Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, Bethesda, MD 20892.
Organizational Affiliation: