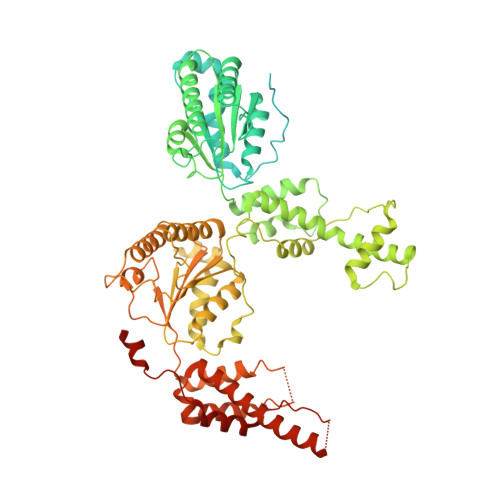
Cryo-EM structure of the essential ribosome assembly AAA-ATPase Rix7.
Lo, Y.H., Sobhany, M., Hsu, A.L., Ford, B.L., Krahn, J.M., Borgnia, M.J., Stanley, R.E.(2019) Nat Commun 10: 513-513
- PubMed: 30705282
- DOI: https://doi.org/10.1038/s41467-019-08373-0
- Primary Citation of Related Structures:
6MAT - PubMed Abstract:
Rix7 is an essential type II AAA-ATPase required for the formation of the large ribosomal subunit. Rix7 has been proposed to utilize the power of ATP hydrolysis to drive the removal of assembly factors from pre-60S particles, but the mechanism of release is unknown. Rix7's mammalian homolog, NVL2 has been linked to cancer and mental illness disorders, highlighting the need to understand the molecular mechanisms of this essential machine. Here we report the cryo-EM reconstruction of the tandem AAA domains of Rix7 which form an asymmetric stacked homohexameric ring. We trapped Rix7 with a polypeptide in the central channel, revealing Rix7's role as a molecular unfoldase. The structure establishes that type II AAA-ATPases lacking the aromatic-hydrophobic motif within the first AAA domain can engage a substrate throughout the entire central channel. The structure also reveals that Rix7 contains unique post-α7 insertions within both AAA domains important for Rix7 function.
- Signal Transduction Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Department of Health and Human Services, 111T. W. Alexander Drive, Research Triangle Park, NC, 27709, USA.
Organizational Affiliation: