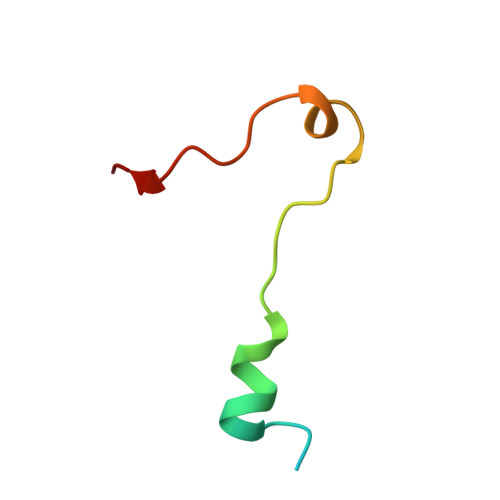
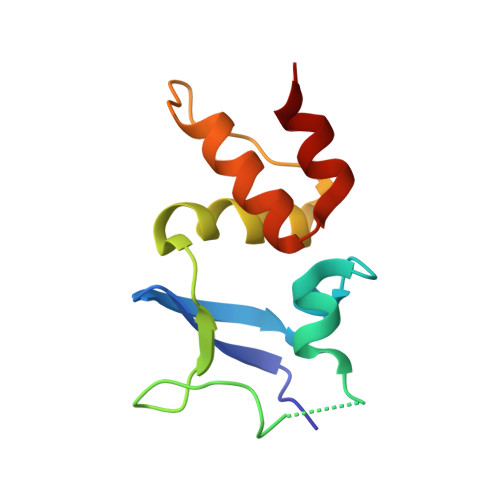Structural basis for KCTD-mediated rapid desensitization of GABABsignalling.
Zheng, S., Abreu, N., Levitz, J., Kruse, A.C.(2019) Nature 567: 127-131
- PubMed: 30814734
- DOI: https://doi.org/10.1038/s41586-019-0990-0
- Primary Citation of Related Structures:
6M8R, 6M8S - PubMed Abstract:
The GABA B (γ-aminobutyric acid type B) receptor is one of the principal inhibitory neurotransmitter receptors in the brain, and it signals through heterotrimeric G proteins to activate a variety of effectors, including G-protein-coupled inwardly rectifying potassium channels (GIRKs) 1,2 . GABA B -receptor signalling is tightly regulated by auxiliary subunits called KCTDs, which control the kinetics of GIRK activation and desensitization 3-5 . However, the mechanistic basis for KCTD modulation of GABA B signalling remains incompletely understood. Here, using a combination of X-ray crystallography, electron microscopy, and functional and biochemical experiments, we reveal the molecular details of KCTD binding to both GABA B receptors and G-protein βγ subunits. KCTDs associate with the receptor by forming an asymmetric pentameric ring around a region of the receptor carboxy-terminal tail, while a second KCTD domain, H1, engages in a symmetric interaction with five copies of Gβγ in which the G-protein subunits also interact directly with one another. We further show that KCTD binding to Gβγ is highly cooperative, defining a model in which KCTD proteins cooperatively strip G proteins from GIRK channels to induce rapid desensitization following receptor activation. These results provide a framework for understanding the molecular basis for the precise temporal control of GABA B signalling by KCTD proteins.
- Department of Biological Chemistry and Molecular Pharmacology, Blavatnik Institute, Harvard Medical School, Boston, MA, USA.
Organizational Affiliation: