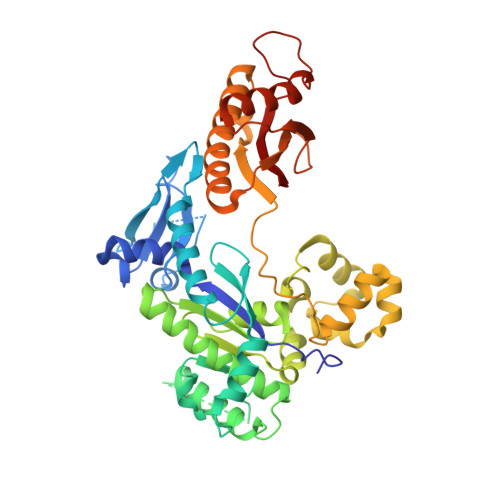
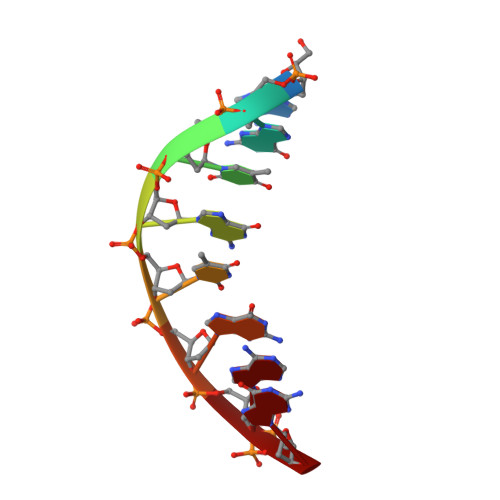
Bypassing a 8,5'-cyclo-2'-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution.
Weng, P.J., Gao, Y., Gregory, M.T., Wang, P., Wang, Y., Yang, W.(2018) Proc Natl Acad Sci U S A 115: 10660-10665
- PubMed: 30275308
- DOI: https://doi.org/10.1073/pnas.1812856115
- Primary Citation of Related Structures:
6M7O, 6M7P, 6M7T, 6M7U, 6M7V - PubMed Abstract:
Oxidatively induced DNA lesions 8,5'-cyclopurine-2'-deoxynucleosides (cdPus) are prevalent and cytotoxic by impeding DNA replication and transcription. Both the 5' R - and 5' S -diastereomers of cdPu can be removed by nucleotide excision repair; however, the 5' S -cdPu is more resistant to repair than the 5' R counterpart. Here, we report the crystal structures of human polymerase (Pol) η bypassing 5' S -8,5'-cyclo-2'-deoxyadenosine (cdA) in insertion and the following two extension steps. The cdA-containing DNA structures vary in response to the protein environment. Supported by the "molecular splint" of Pol η, the structure of 5' S -cdA at 1.75-Å resolution reveals that the backbone is pinched toward the minor groove and the adenine base is tilted. In the templating position, the cdA takes up the extra space usually reserved for the thymine dimer, and dTTP is efficiently incorporated by Pol η in the presence of Mn 2+ Rigid distortions of the DNA duplex by cdA, however, prevent normal base pairing and hinder immediate primer extension by Pol η. Our results provide structural insights into the strong replication blockage effect and the mutagenic property of the cdPu lesions in cells.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892.
Organizational Affiliation: