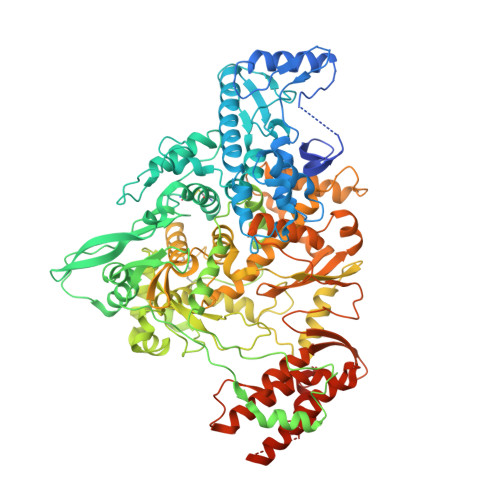
Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., Wang, T., Sun, Q., Ming, Z., Zhang, L., Ge, J., Zheng, L., Zhang, Y., Wang, H., Zhu, Y., Zhu, C., Hu, T., Hua, T., Zhang, B., Yang, X., Li, J., Yang, H., Liu, Z., Xu, W., Guddat, L.W., Wang, Q., Lou, Z., Rao, Z.(2020) Science 368: 779-782
- PubMed: 32277040
- DOI: https://doi.org/10.1126/science.abb7498
- Primary Citation of Related Structures:
6M71, 7BTF - PubMed Abstract:
A novel coronavirus [severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2)] outbreak has caused a global coronavirus disease 2019 (COVID-19) pandemic, resulting in tens of thousands of infections and thousands of deaths worldwide. The RNA-dependent RNA polymerase [(RdRp), also named nsp12] is the central component of coronaviral replication and transcription machinery, and it appears to be a primary target for the antiviral drug remdesivir. We report the cryo-electron microscopy structure of COVID-19 virus full-length nsp12 in complex with cofactors nsp7 and nsp8 at 2.9-angstrom resolution. In addition to the conserved architecture of the polymerase core of the viral polymerase family, nsp12 possesses a newly identified β-hairpin domain at its N terminus. A comparative analysis model shows how remdesivir binds to this polymerase. The structure provides a basis for the design of new antiviral therapeutics that target viral RdRp.
- Laboratory of Structural Biology, School of Life Sciences, and School of Medicine, Tsinghua University, Beijing, China.
Organizational Affiliation: