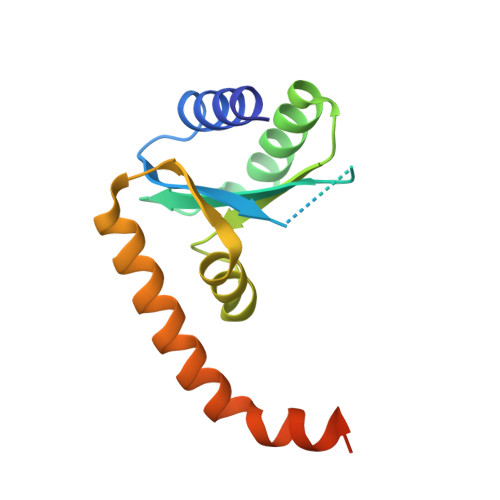
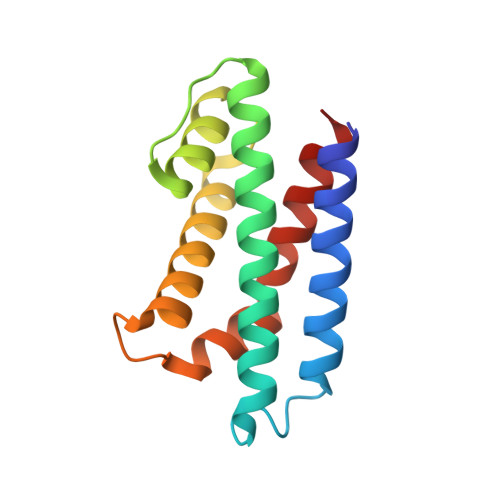
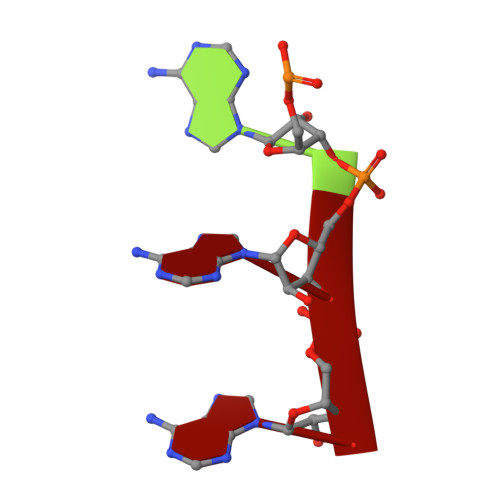
Novel polyadenylylation-dependent neutralization mechanism of the HEPN/MNT toxin/antitoxin system.
Yao, J., Zhen, X., Tang, K., Liu, T., Xu, X., Chen, Z., Guo, Y., Liu, X., Wood, T.K., Ouyang, S., Wang, X.(2020) Nucleic Acids Res 48: 11054-11067
- PubMed: 33045733
- DOI: https://doi.org/10.1093/nar/gkaa855
- Primary Citation of Related Structures:
6M6U, 6M6V, 6M6W, 7BXO - PubMed Abstract:
The two-gene module HEPN/MNT is predicted to be the most abundant toxin/antitoxin (TA) system in prokaryotes. However, its physiological function and neutralization mechanism remains obscure. Here, we discovered that the MntA antitoxin (MNT-domain protein) acts as an adenylyltransferase and chemically modifies the HepT toxin (HEPN-domain protein) to block its toxicity as an RNase. Biochemical and structural studies revealed that MntA mediates the transfer of three AMPs to a tyrosine residue next to the RNase domain of HepT in Shewanella oneidensis. Furthermore, in vitro enzymatic assays showed that the three AMPs are transferred to HepT by MntA consecutively with ATP serving as the substrate, and this polyadenylylation is crucial for reducing HepT toxicity. Additionally, the GSX10DXD motif, which is conserved among MntA proteins, is the key active motif for polyadenylylating and neutralizing HepT. Thus, HepT/MntA represents a new type of TA system, and the polyadenylylation-dependent TA neutralization mechanism is prevalent in bacteria and archaea.
- Key Laboratory of Tropical Marine Bio-resources and Ecology, Guangdong Key Laboratory of Marine Materia Medica, Innovation Academy of South China Sea Ecology and Environmental Engineering, South China Sea Institute of Oceanology, Chinese Academy of Sciences, 164 West Xingang Road, Guangzhou 510301, China.
Organizational Affiliation: