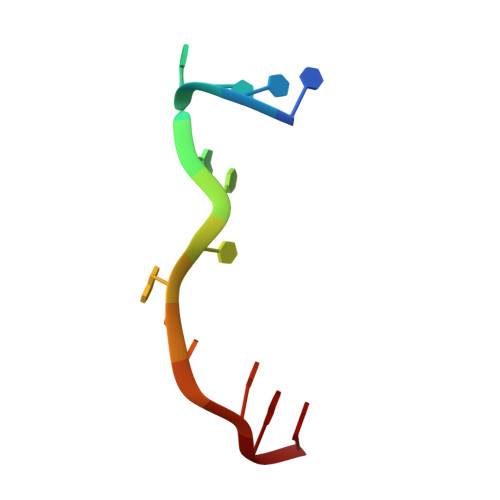
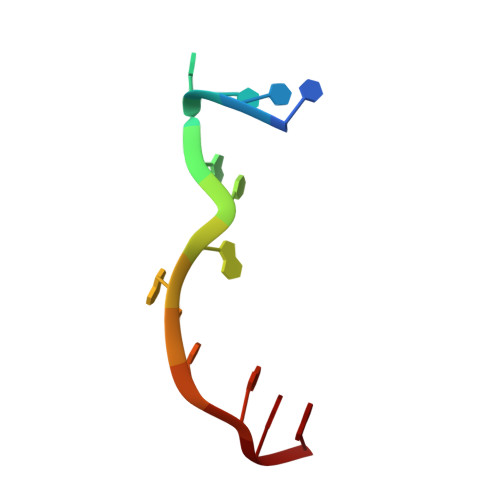Structural Basis for Targeting T:T Mismatch with Triaminotriazine-Acridine Conjugate Induces a U-Shaped Head-to-Head Four-Way Junction in CTG Repeat DNA.
Chien, C.M., Wu, P.C., Satange, R., Chang, C.C., Lai, Z.L., Hagler, L.D., Zimmerman, S.C., Hou, M.H.(2020) J Am Chem Soc 142: 11165-11172
- PubMed: 32478511
- DOI: https://doi.org/10.1021/jacs.0c03591
- Primary Citation of Related Structures:
6M4T, 6M5J - PubMed Abstract:
The potent DNA-binding compound triaminotriazine-acridine conjugate (Z1) functions by targeting T:T mismatches in CTG trinucleotide repeats that are responsible for causing neurological diseases such as myotonic dystrophy type 1, but its binding mechanism remains unclear. We solved a crystal structure of Z1 in a complex with DNA containing three consecutive CTG repeats with three T:T mismatches. Crystallographic studies revealed that direct intercalation of two Z1 molecules at both ends of the CTG repeat induces thymine base flipping and DNA backbone deformation to form a four-way junction. The core of the complex unexpectedly adopts a U-shaped head-to-head topology to form a crossover of each chain at the junction site. The crossover junction is held together by two stacked G:C pairs at the central core that rotate with respect to each other in an X-shape to form two nonplanar minor-groove-aligned G·C·G·C tetrads. Two stacked G:C pairs on both sides of the center core are involved in the formation of pseudo-continuous duplex DNA. Four metal-mediated base pairs are observed between the N7 atoms of G and Co II , an interaction that strongly preserves the central junction site. Beyond revealing a new type of ligand-induced, four-way junction, these observations enhance our understanding of the specific supramolecular chemistry of Z1 that is essential for the formation of a noncanonical DNA superstructure. The structural features described here serve as a foundation for the design of new sequence-specific ligands targeting mismatches in the repeat-associated structures.
- Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, United States.
Organizational Affiliation: