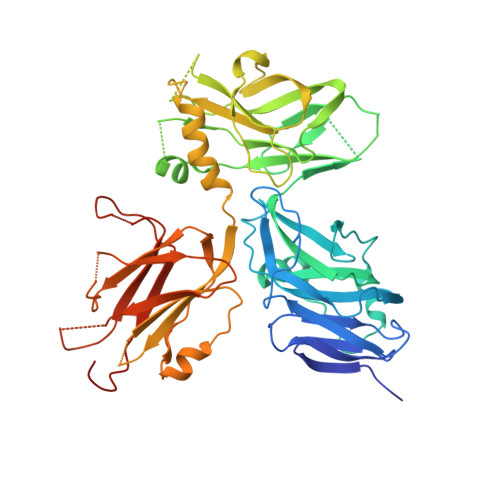
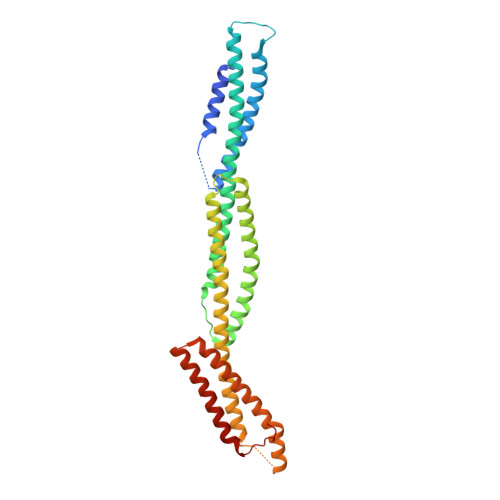
Structural Basis Underlying Strong Interactions between Ankyrins and Spectrins.
Li, J., Chen, K., Zhu, R., Zhang, M.(2020) J Mol Biology 432: 3838-3850
- PubMed: 32353364
- DOI: https://doi.org/10.1016/j.jmb.2020.04.023
- Primary Citation of Related Structures:
6M3P, 6M3Q, 6M3R - PubMed Abstract:
Ankyrins (encoded by ANK1/2/3 corresponding to Ankyrin-R/B/G or AnkR/B/G), via binding to spectrins, connect plasma membranes with actin cytoskeleton to maintain mechanical strengths and to modulate excitabilities of diverse cells such as neurons, muscle cells, and erythrocytes. Cellular and genetic evidences suggest that each isoform of ankyrins pairs with a specific β-spectrin in discrete subcellular membrane microdomains for distinct functions, although the molecular mechanisms underlying such ankyrin/β-spectrin pairings are unknown. In this study, we discover that a conserved and short extension N-terminal to the ZU5 N -ZU5 C -UPA tandem (exZZU) is critical for each ankyrin to bind to β-spectrins with high affinities. Structures of AnkB/G exZZU in complex with spectrin repeats13-15 of β2/β4-spectrins solved here reveal that the extension sequence of exZZU forms an additional β-strand contributing to the structural stability and enhanced affinity of each ZU5 N /spectrin repeat interaction. The complex structures further reveal that the UPA domain of exZZU directly participates in spectrin binding. Formation of the exZZU supramodule juxtaposes the ZU5 N and UPA domains for simultaneous interacting with spectrin repeats 14 and 15. However, our biochemical and structural investigations indicate that the direct and strong interactions between ankyrins and β-spectrins do not appear to determine their pairing specificities. Therefore, there likely exists additional mechanism(s) for modulating functional pairings between ankyrins and β-spectrins in cells.
- Division of Life Science, State Key Laboratory of Molecular Neuroscience, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong, China; Division of Cell, Developmental and Integrative Biology, School of Medicine, South China University of Technology, Guangzhou 510006, China.
Organizational Affiliation: