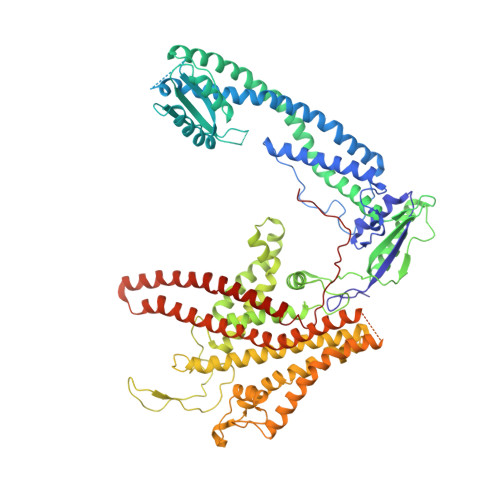
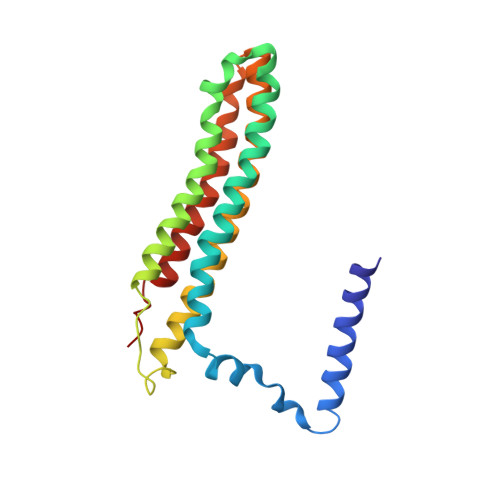
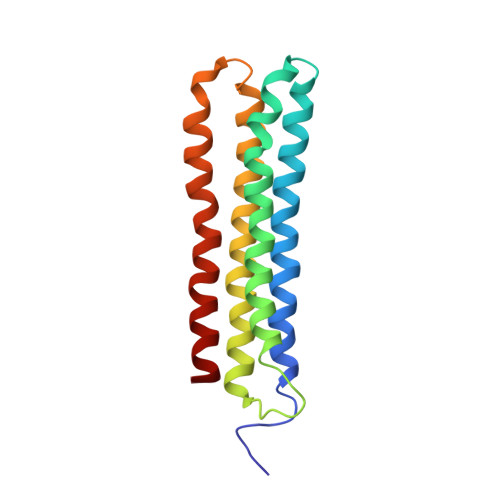
Cryo-EM and MD infer water-mediated proton transport and autoinhibition mechanisms of V o complex.
Roh, S.H., Shekhar, M., Pintilie, G., Chipot, C., Wilkens, S., Singharoy, A., Chiu, W.(2020) Sci Adv 6
- PubMed: 33028525
- DOI: https://doi.org/10.1126/sciadv.abb9605
- Primary Citation of Related Structures:
6M0R, 6M0S - PubMed Abstract:
Rotary vacuolar adenosine triphosphatases (V-ATPases) drive transmembrane proton transport through a V o proton channel subcomplex. Despite recent high-resolution structures of several rotary ATPases, the dynamic mechanism of proton pumping remains elusive. Here, we determined a 2.7-Å cryo-electron microscopy (cryo-EM) structure of yeast V o proton channel in nanodisc that reveals the location of ordered water molecules along the proton path, details of specific protein-lipid interactions, and the architecture of the membrane scaffold protein. Moreover, we uncover a state of V o that shows the c -ring rotated by ~14°. Molecular dynamics simulations demonstrate that the two rotary states are in thermal equilibrium and depict how the protonation state of essential glutamic acid residues couples water-mediated proton transfer with c -ring rotation. Our cryo-EM models and simulations also rationalize a mechanism for inhibition of passive proton transport as observed for free V o that is generated as a result of V-ATPase regulation by reversible disassembly in vivo.
- School of Biological Sciences, Seoul National University, Seoul 08826, South Korea. wahc@stanford.edu shroh@snu.ac.kr asinghar@asu.edu wilkenss@upstate.edu.
Organizational Affiliation: