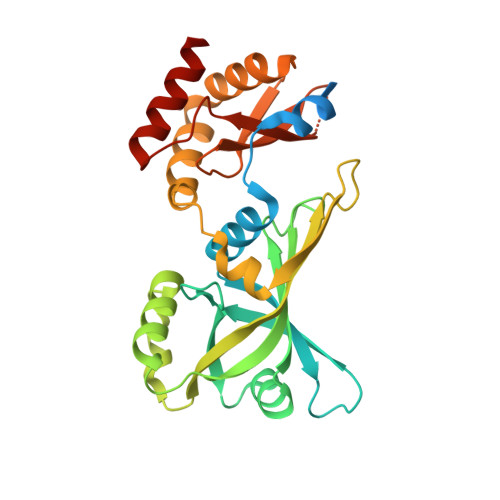
The Arabidopsis locus AT3G03890 encodes a dimeric beta-barrel protein implicated in heme degradation.
Wang, J., Guo, Q., Li, X., Wang, X., Liu, L.(2020) Biochem J
- PubMed: 33284325
- DOI: https://doi.org/10.1042/BCJ20200712
- Primary Citation of Related Structures:
6M09, 6M0A - PubMed Abstract:
Plant tetrapyrroles, including heme and bilins, are synthesized in plastids. Heme oxygenase (HO) catalyzes the oxidative cleavage of heme to the linear tetrapyrrole biliverdin as the initial step in bilin biosynthesis. Besides the canonical α-helical HO that is conserved from prokaryotes to human, a subfamily of non-canonical dimeric β-barrel HO has been found in bacteria. In this work, we discovered that the Arabidopsis locus AT3G03890 encodes a dimeric β-barrel protein that is structurally related to the putative non-canonical HO and is located in chloroplasts. The recombinant protein was able to bind and degrade heme in a manner different from known HO proteins. Crystal structure of the heme-protein complex reveals that the heme-binding site is in the interdimer interface and the heme iron is coordinated by a fixed water molecule. Our results identify a new protein that may function additionally in the tetrapyrrole biosynthetic pathway.
- IBCAS, Beijing, China.
Organizational Affiliation: