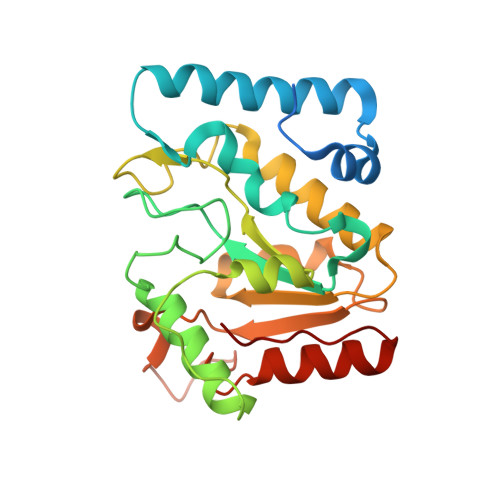
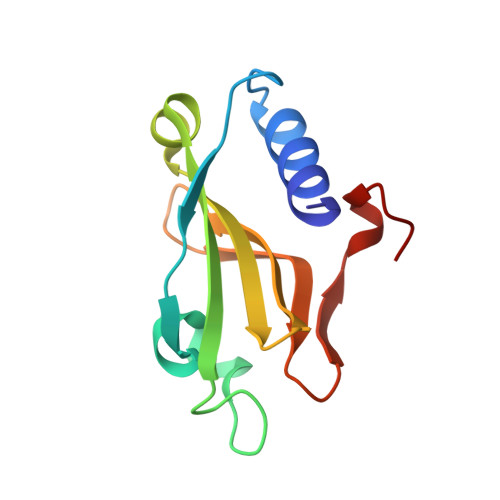
Structural insight into the differential interactions between the DNA mimic protein SAUGI and two gamma herpesvirus uracil-DNA glycosylases.
Liao, Y.T., Lin, S.J., Ko, T.P., Liu, C.Y., Hsu, K.C., Wang, H.C.(2020) Int J Biol Macromol 160: 903-914
- PubMed: 32502608
- DOI: https://doi.org/10.1016/j.ijbiomac.2020.05.267
- Primary Citation of Related Structures:
6LYJ, 6LYV - PubMed Abstract:
Uracil-DNA glycosylases (UDGs) are conserved DNA-repair enzymes that can be found in many species, including herpesviruses. Since they play crucial roles for efficient viral DNA replication in herpesviruses, they have been considered as potential antiviral targets. In our previous work, Staphylococcus aureus SAUGI was identified as a DNA mimic protein that targets UDGs from S. aureus, human, Herpes simplex virus (HSV) and Epstein-Barr virus (EBV). Interestingly, SAUGI has the strongest inhibitory effects with EBVUDG. Here, we determined complex structures of SAUGI with EBVUDG and another γ-herpesvirus UDG from Kaposi's sarcoma-associated herpesvirus (KSHVUDG), which SAUGI fails to effectively inhibit. Structural analysis of the SAUGI/EBVUDG complex suggests that the additional interaction between SAUGI and the leucine loop may explain why SAUGI shows the highest binding capacity with EBVUDG. In contrast, SAUGI appears to make only partial contacts with the key components responsible for the compression and stabilization of the DNA backbone in the leucine loop extension of KSHVUDG. The findings in this study provide a molecular explanation for the differential inhibitory effects and binding strengths that SAUGI has on these two UDGs, and the structural basis of the differences should be helpful in developing inhibitors that would interfere with viral DNA replication.
- The PhD Program for Translational Medicine, College of Medical Science and Technology, Taipei Medical University and Academia Sinica, Taipei 115, Taiwan; Graduate Institute of Translational Medicine, College of Medical Science and Technology, Taipei Medical University, Taipei 110, Taiwan. Electronic address: d622105001@tmu.edu.tw.
Organizational Affiliation: