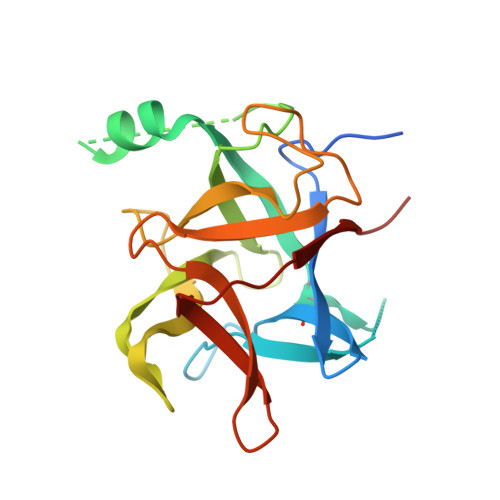
Crystal structure of the N-terminal domain of ryanodine receptor from the honeybee, Apis mellifera.
Zhou, Y., Wang, W., Salauddin, N.M., Lin, L., You, M., You, S., Yuchi, Z.(2020) Insect Biochem Mol Biol 125: 103454-103454
- PubMed: 32781205
- DOI: https://doi.org/10.1016/j.ibmb.2020.103454
- Primary Citation of Related Structures:
6LQK - PubMed Abstract:
Ryanodine receptors (RyRs) are the molecular target of diamides, a new chemical class of insecticides. Diamide insecticides are used to control lepidopteran pests and were considered relatively safe for mammals and non-targeted beneficial insects, including honey bees. However, recent studies showed that exposure to diamides could cause long-lasting locomotor deficits of bees. Here we report the crystal structure of RyR N-terminal domain A (NTD-A) from the honeybee, Apis mellifera, at 2.5 Å resolution. It shows a similar overall fold as the RyR NTD-A from mammals and the diamondback moth (DBM), Plutella xylostella, and still several loops located at the inter-domain interfaces show insect-specific or bee-specific structural features. A potential insecticide-binding pocket formed by loop9 and loop13 is conserved in lepidopteran but different in both mammals and bees, making it a good candidate targeting site for the development of pest-selective insecticides. Furthermore, a conserved intra-domain disulfide bond was observed in both DBM and bee RyR NTD-A crystal structures, which explains their higher thermal stability compared to mammalian RyR NTD-A. This work provides a basis for the development of novel insecticides with better selectivity between pests and bees by targeting a distinct site on pest RyRs, which would be a promising strategy to overcome the current toxicity problem.
- Tianjin Key Laboratory for Modern Drug Delivery & High-Efficiency, Collaborative Innovation Center of Chemical Science and Engineering, School of Pharmaceutical Science and Technology, Tianjin University, Tianjin, 300072, China; State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Institute of Applied Ecology, Fujian Agriculture and Forestry University, Fuzhou, 350002, China; Joint International Research Laboratory of Ecological Pest Control, Ministry of Education, Fuzhou, 350002, China; Key Laboratory of Integrated Pest Management for Fujian-Taiwan Crops, Ministry of Agriculture, Fuzhou, 350002, China.
Organizational Affiliation: