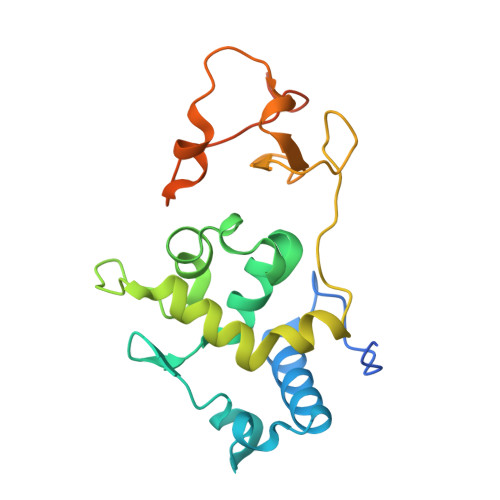
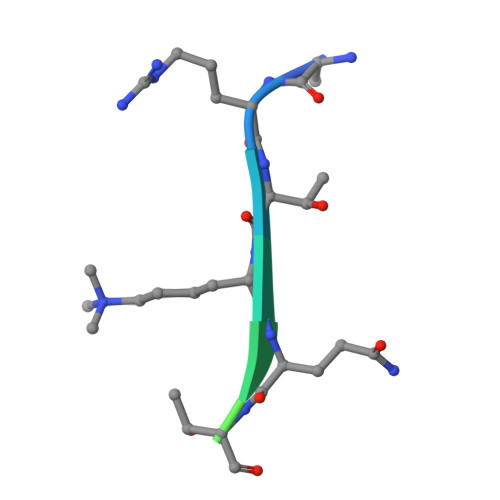
Dual Recognition of H3K4me3 and DNA by the ISWI Component ARID5 Regulates the Floral Transition in Arabidopsis.
Tan, L.M., Liu, R., Gu, B.W., Zhang, C.J., Luo, J., Guo, J., Wang, Y., Chen, L., Du, X., Li, S., Shao, C.R., Su, Y.N., Cai, X.W., Lin, R.N., Li, L., Chen, S., Du, J., He, X.J.(2020) Plant Cell 32: 2178-2195
- PubMed: 32358072
- DOI: https://doi.org/10.1105/tpc.19.00944
- Primary Citation of Related Structures:
6LQE, 6LQF - PubMed Abstract:
Chromatin remodeling and histone modifications are important for development and floral transition in plants. However, it is largely unknown whether and how these two epigenetic regulators coordinately regulate the important biological processes. Here, we identified three types of Imitation Switch (ISWI) chromatin-remodeling complexes in Arabidopsis ( Arabidopsis thaliana ). We found that AT-RICH INTERACTING DOMAIN5 (ARID5), a subunit of a plant-specific ISWI complex, can regulate development and floral transition. The ARID-PHD dual domain cassette of ARID5 recognizes both the H3K4me3 histone mark and AT-rich DNA. We determined the ternary complex structure of the ARID5 ARID-PHD cassette with an H3K4me3 peptide and an AT-containing DNA. The H3K4me3 peptide is combinatorially recognized by the PHD and ARID domains, while the DNA is specifically recognized by the ARID domain. Both PHD and ARID domains are necessary for the association of ARID5 with chromatin. The results suggest that the dual recognition of AT-rich DNA and H3K4me3 by the ARID5 ARID-PHD cassette may facilitate the association of the ISWI complex with specific chromatin regions to regulate development and floral transition.
- National Institute of Biological Sciences, Beijing 102206, China.
Organizational Affiliation: