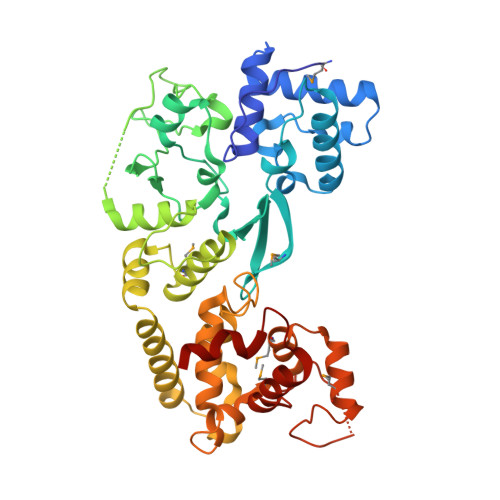
Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Wang, B., Xu, W., Yang, H.(2020) Cell Res 30: 185-187
- PubMed: 31913359
- DOI: https://doi.org/10.1038/s41422-020-0274-0
- Primary Citation of Related Structures:
6LNB, 6LNC, 6LND - State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai, 200031, China.
Organizational Affiliation: