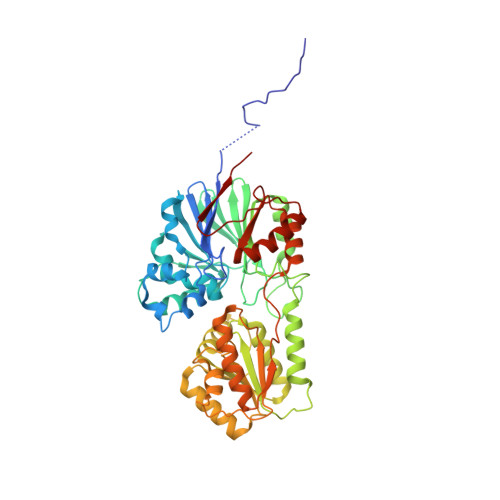
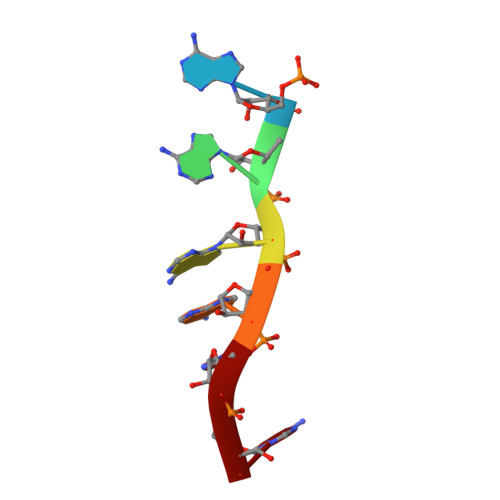
A newly identified duplex RNA unwinding activity of archaeal RNase J depends on processive exoribonucleolysis coupled steric occlusion by its structural archaeal loops.
Li, J., Hou, Y., Gu, X., Yue, L., Guo, L., Li, D., Dong, X.(2020) RNA Biol 17: 1480-1491
- PubMed: 32552320
- DOI: https://doi.org/10.1080/15476286.2020.1777379
- Primary Citation of Related Structures:
6LLB - PubMed Abstract:
RNase J is a prokaryotic 5'-3' exo/endoribonuclease that functions in mRNA decay and rRNA maturation. Here, we report a novel duplex unwinding activity of mpy-RNase J, an archaeal RNase J from Methanolobus psychrophilus , which enables it to degrade duplex RNAs with hairpins up to 40 bp when linking a 5' single-stranded overhangs of ≥ 7 nt, corresponding to the RNA channel length. A 6-nt RNA-mpy-RNase J-S247A structure reveals the RNA-interacting residues and a steric barrier at the RNA channel entrance comprising two archaeal loops and two helices. Mutagenesis of the residues key to either exoribonucleolysis or RNA translocation diminished the duplex unwinding activity. Substitution of the residues in the steric barrier yielded stalled degradation intermediates at the duplex RNA regions. Thus, an exoribonucleolysis-driven and steric occlusion-based duplex unwinding mechanism was identified. The duplex unwinding activity confers mpy-RNase J the capability of degrading highly structured RNAs, including the bacterial REP RNA, and archaeal mRNAs, rRNAs, tRNAs, SRPs, RNase P and CD-box RNAs, providing an indicative of the potential key roles of mpy-RNase J in pleiotropic RNA metabolisms. Hydrolysis-coupled duplex unwinding activity was also detected in a bacterial RNase J, which may use a shared but slightly different unwinding mechanism from archaeal RNase Js, indicating that duplex unwinding is a common property of the prokaryotic RNase Js.
- State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences , Beijing, PR China.
Organizational Affiliation: