Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Chiu, Y.C., Tseng, M.C., Hsu, C.H.(2021) ACS Catal 11: 11075-11090
Experimental Data Snapshot
Starting Model: experimental
View more details
(2021) ACS Catal 11: 11075-11090
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ADP-ribose 1''-phosphate phosphatase | 180 | Saccharomyces cerevisiae S288C | Mutation(s): 1 Gene Names: POA1, YBR022W, YBR0304 EC: 3.1.3.84 (PDB Primary Data), 3.2.2 (PDB Primary Data) | 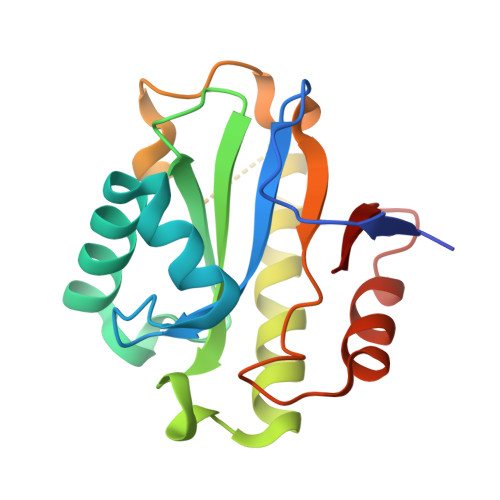 | |
UniProt | |||||
Find proteins for P38218 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P38218 Go to UniProtKB: P38218 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P38218 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| APR (Subject of Investigation/LOI) Query on APR | C [auth A] | ADENOSINE-5-DIPHOSPHORIBOSE C15 H23 N5 O14 P2 SRNWOUGRCWSEMX-KEOHHSTQSA-N |  | ||
| ACT Query on ACT | B [auth A] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 82.218 | α = 90 |
| b = 82.218 | β = 90 |
| c = 52.416 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministry of Science and Technology (Taiwan) | Taiwan | 108-2628-B-002-013 |
| Ministry of Science and Technology (Taiwan) | Taiwan | 108-2113-M-002-011 |