Crystal structure of YggS family pyridoxal phosphate-dependent enzyme PipY from Fusobacterium nucleatum
Chen, Y., Wang, L., Shang, F., Lan, J., Liu, W., Xu, Y.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Pyridoxal phosphate homeostasis protein | 223 | Fusobacterium nucleatum subsp. nucleatum | Mutation(s): 2 Gene Names: RO03_02185 | 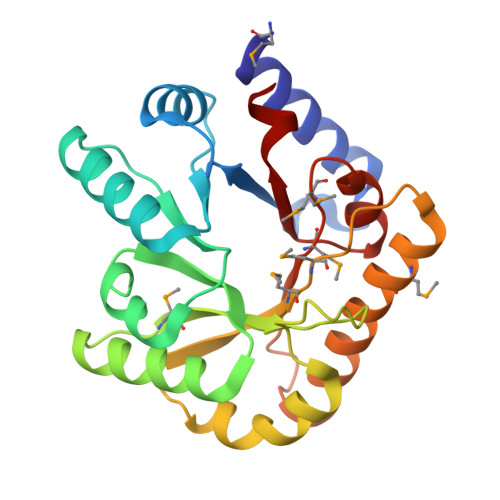 | |
UniProt | |||||
Find proteins for Q8RFW9 (Fusobacterium nucleatum subsp. nucleatum (strain ATCC 25586 / DSM 15643 / BCRC 10681 / CIP 101130 / JCM 8532 / KCTC 2640 / LMG 13131 / VPI 4355)) Explore Q8RFW9 Go to UniProtKB: Q8RFW9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8RFW9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PO4 (Subject of Investigation/LOI) Query on PO4 | D [auth A], E [auth B], F [auth C] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B, C | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 37.929 | α = 90 |
| b = 146.375 | β = 93.355 |
| c = 74.128 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China | China | 31200556 |
| National Natural Science Foundation of China | China | 21272031 |