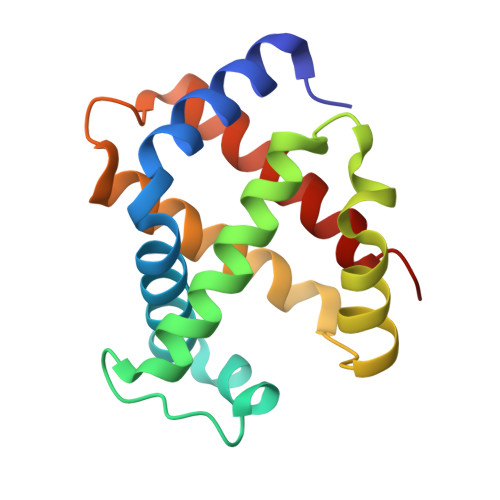
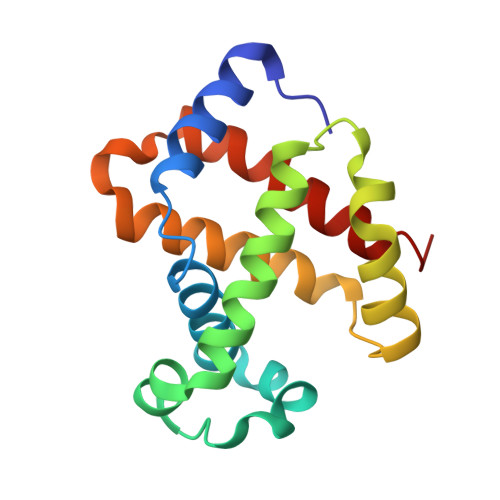
Genetically engineered haemoglobin wrapped covalently with human serum albumins as an artificial O2carrier.
Funaki, R., Okamoto, W., Endo, C., Morita, Y., Kihira, K., Komatsu, T.(2020) J Mater Chem B 8: 1139-1145
- PubMed: 31840728
- DOI: https://doi.org/10.1039/c9tb02184a
- Primary Citation of Related Structures:
6KYE - PubMed Abstract:
We describe the synthesis and O 2 affinity of genetically engineered human adult haemoglobin (rHbA) wrapped covalently with recombinant human serum albumins (rHSAs) as an artificial O 2 carrier used for a completely synthetic red blood cell (RBC) substitute. Wild-type rHbA [rHbA(wt)] expressed in yeast species Pichia pastoris shows an identical amino acid sequence and three-dimensional structure to those of native HbA. It is particularly interesting that two orientations of the prosthetic haem group in rHbA(wt) were aligned by gentle heating in the natural form. Covalent wrapping of rHbA(wt) with three rHSAs conferred a core-shell structured haemoglobin-albumin cluster: rHbA(wt)-rHSA 3 . Three variant clusters containing an rHbA mutant core were also created: Leu-β28 → Phe, Leu-β28 → Trp, and Leu-β28 → Tyr/His-β63 → Gln. Replacement of Leu-β28 with Trp decreased the distal space in the haem pocket, thereby yielding a cluster with moderately low O 2 affinity which is nearly the same as that of human RBC.
- Department of Applied Chemistry, Faculty of Science and Engineering, Chuo University, 1-13-27 Kasuga, Bunkyo-ku, Tokyo 112-8551, Japan. komatsu@kc.chuo-u.ac.jp.
Organizational Affiliation: