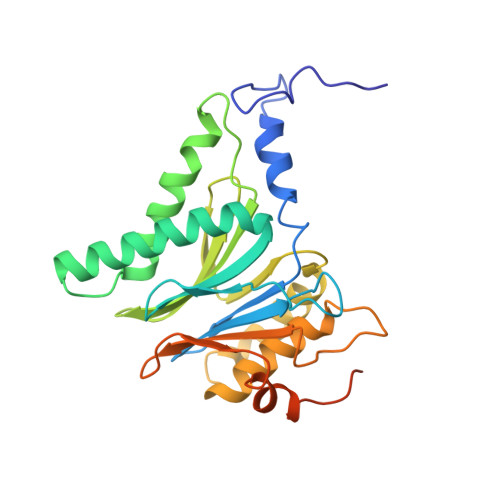
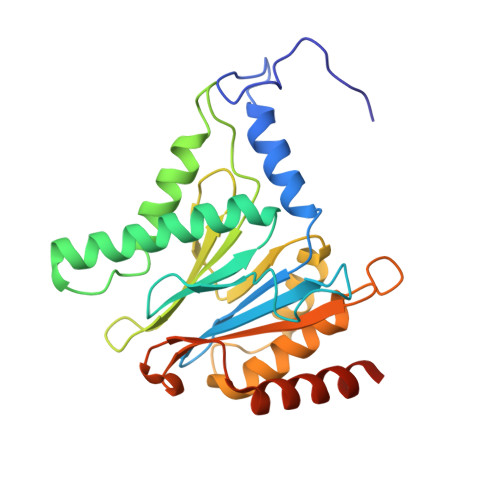
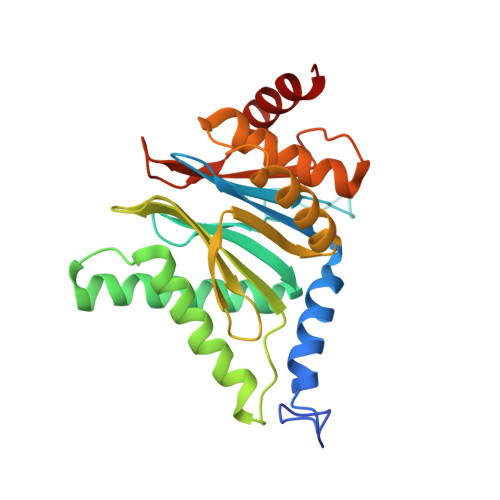
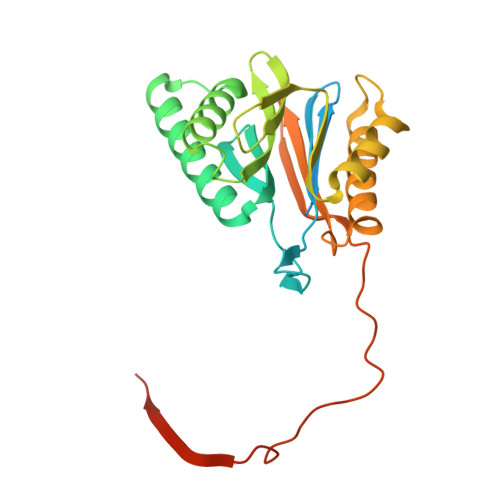
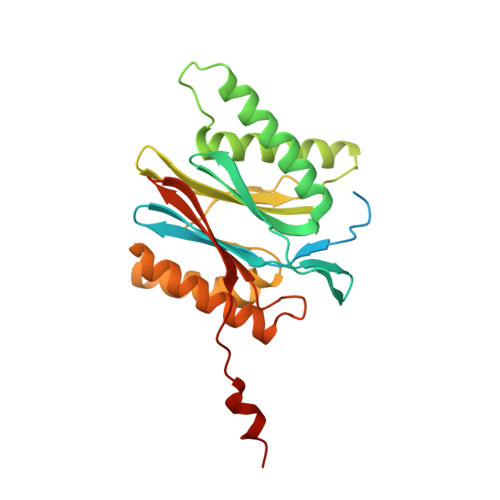
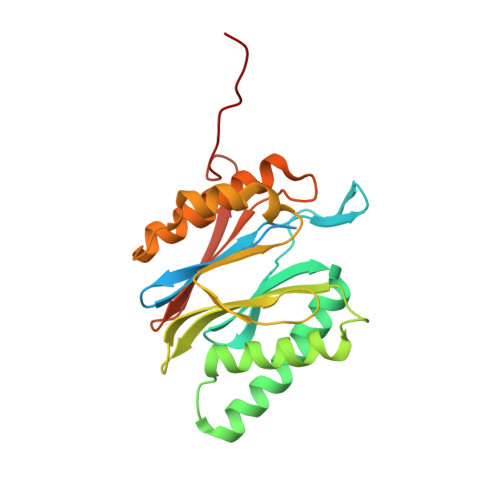
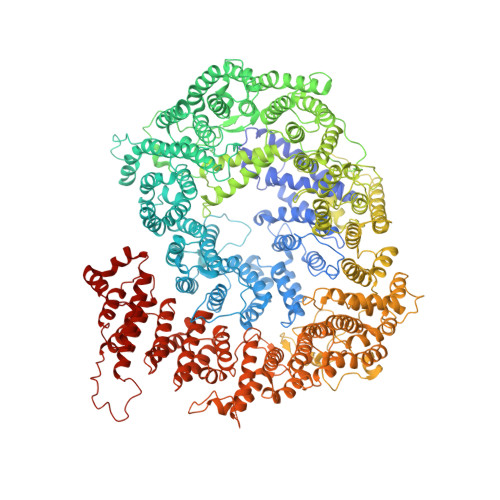
Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures.
Guan, H., Wang, Y., Yu, T., Huang, Y., Li, M., Saeed, A.F.U.H., Perculija, V., Li, D., Xiao, J., Wang, D., Zhu, P., Ouyang, S.(2020) PLoS Biol 18: e3000654-e3000654
- PubMed: 32134919
- DOI: https://doi.org/10.1371/journal.pbio.3000654
- Primary Citation of Related Structures:
6KWX, 6KWY - PubMed Abstract:
Proteasomes are highly abundant and conserved protease complexes that eliminate unwanted proteins in the cells. As a single-chain ATP-independent nuclear proteasome activator, proteasome activator 200 (PA200) associates with 20S core particle to form proteasome complex that catalyzes polyubiquitin-independent degradation of acetylated histones, thus playing a pivotal role in DNA repair and spermatogenesis. Here, we present cryo-electron microscopy (cryo-EM) structures of the human PA200-20S complex and PA200 at 2.72 Å and 3.75 Å, respectively. PA200 exhibits a dome-like architecture that caps 20S and uses its C-terminal YYA (Tyr-Tyr-Ala) to induce the α-ring rearrangements and partial opening of the 20S gate. Our structural data also indicate that PA200 has two openings formed by numerous positively charged residues that respectively bind (5,6)-bisdiphosphoinositol tetrakisphosphate (5,6[PP]2-InsP4) and inositol hexakisphosphate (InsP6) and are likely to be the gates that lead unfolded proteins through PA200 and into the 20S. Besides, our structural analysis of PA200 found that the bromodomain (BRD)-like (BRDL) domain of PA200 shows considerable sequence variation in comparison to other human BRDs, as it contains only 82 residues because of a short ZA loop, and cannot be classified into any of the eight typical human BRD families. Taken together, the results obtained from this study provide important insights into human PA200-induced 20S gate opening for substrate degradation and the opportunities to explore the mechanism for its recognition of H4 histone in acetylation-mediated proteasomal degradation.
- The Key Laboratory of Innate Immune Biology of Fujian Province, Provincial University Key Laboratory of Cellular Stress Response and Metabolic Regulation, Biomedical Research Center of South China, Key Laboratory of OptoElectronic Science and Technology for Medicine of Ministry of Education, College of Life Sciences, Fujian Normal University, Fuzhou, China.
Organizational Affiliation: