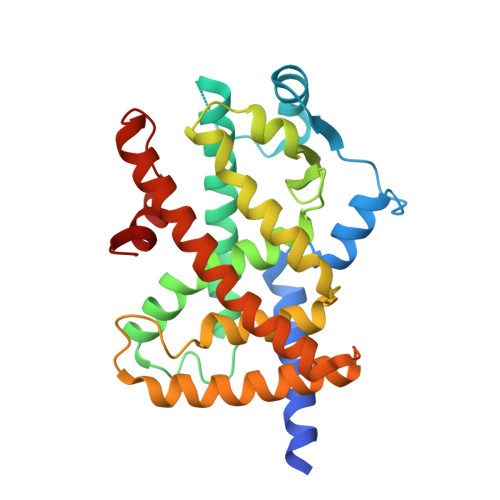
Structural Basis for the Regulation of PPAR gamma Activity by Imatinib.
Jang, J.Y., Kim, H.J., Han, B.W.(2019) Molecules 24
- PubMed: 31581474
- DOI: https://doi.org/10.3390/molecules24193562
- Primary Citation of Related Structures:
6KTM, 6KTN - PubMed Abstract:
Imatinib is an effective anticancer drug for the treatment of leukemia. Interestingly, when an FDA-approved drug library was tested for agents that block peroxisome proliferator-activated receptor γ (PPARγ) phosphorylation at Ser245 to evaluate possibilities of antidiabetic drug repositioning, imatinib was determined as a PPARγ antagonist ligand. However, it is not well understood how imatinib binds to PPARγ or would improve insulin sensitivity without classical agonism. Here, we report the crystal structure of the PPARγ R288A mutant in complex with imatinib. Imatinib bound to Arm2 and Arm3 regions in the ligand-binding domain (LBD) of PPARγ, of which the Arm3 region is closely related to the inhibition of PPARγ phosphorylation at Ser245. The binding of imatinib in LBD induced a stable conformation of helix H2' and the Ω loop compared with the ligand-free state. In contrast, imatinib does not interact with Tyr473 on PPARγ helix H12, which is important for the classical agonism associated with side effects. Our study provides new structural insights into the PPARγ regulation by imatinib and may contribute to the development of new antidiabetic drugs targeting PPARγ while minimizing known side effects.
- Research Institute of Pharmaceutical Sciences, College of Pharmacy, Seoul National University, Seoul 08826, Korea.
Organizational Affiliation: