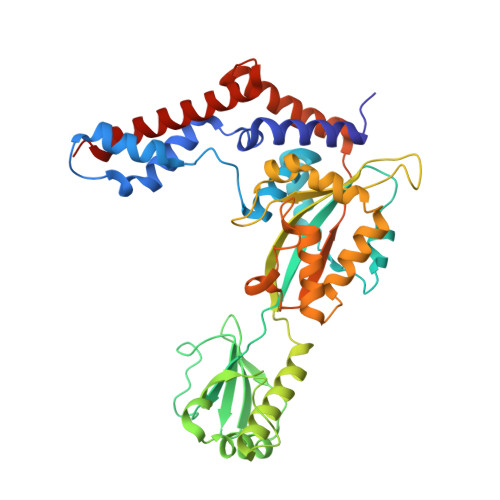
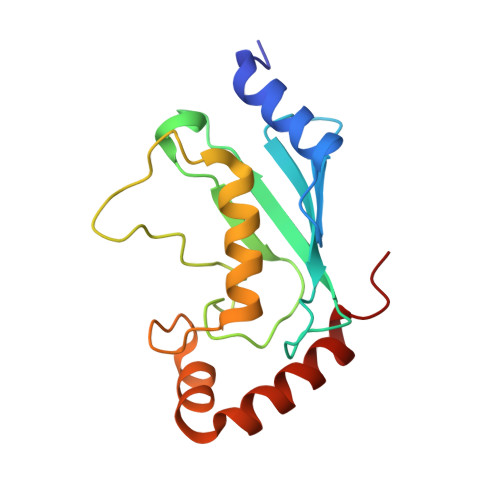
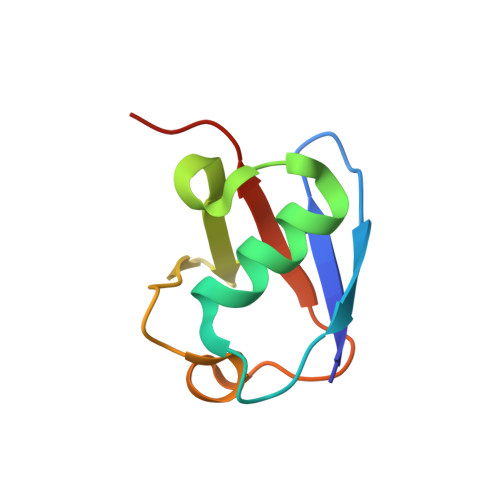
Molecular Basis of Ubiquitination Catalyzed by the Bacterial Transglutaminase MavC.
Guan, H., Fu, J., Yu, T., Wang, Z.X., Gan, N., Huang, Y., Perculija, V., Li, Y., Luo, Z.Q., Ouyang, S.(2020) Adv Sci (Weinh) 7: 2000871-2000871
- PubMed: 32596129
- DOI: https://doi.org/10.1002/advs.202000871
- Primary Citation of Related Structures:
6KL4 - PubMed Abstract:
The Legionella pneumophila effector MavC is a transglutaminase that carries out atypical ubiquitination of the host ubiquitin (Ub)-conjugation enzyme UBE2N by catalyzing the formation of an isopeptide bond between Gln40 Ub and Lys92 UBE2N , which leads to inhibition of signaling in the NF-κB pathway. In the absence of UBE2N, MavC deamidates Ub at Gln40 or catalyzes self-ubiquitination. However, the mechanisms underlying these enzymatic activities of MavC are poorly understood at the molecular level. This study reports the structure of the MavC-UBE2N-Ub ternary complex representing a snapshot of MavC-catalyzed crosslinking of UBE2N and Ub, which reveals the way by which UBE2N-Ub binds to the Insertion and Tail domains of MavC. Based on the structural and experimental data, the catalytic mechanism for the deamidase and transglutaminase activities of MavC is proposed. Finally, by comparing the structures of MavC and MvcA, the homologous protein that reverses MavC-induced UBE2N ubiquitination, several essential regions and two key residues (Trp255 MavC and Phe268 MvcA ) responsible for their respective enzymatic activities are identified. The results provide insights into the mechanisms for substrate recognition and ubiquitination mediated by MavC as well as explanations for the opposite activity of MavC and MvcA in terms of regulation of UBE2N ubiquitination.
- The Key Laboratory of Innate Immune Biology of Fujian Province Provincial University Key Laboratory of Cellular Stress Response and Metabolic Regulation Biomedical Research Center of South China Key Laboratory of OptoElectronic Science and Technology for Medicine of the Ministry of Education College of Life Sciences Fujian Normal University Fuzhou 350117 China.
Organizational Affiliation: