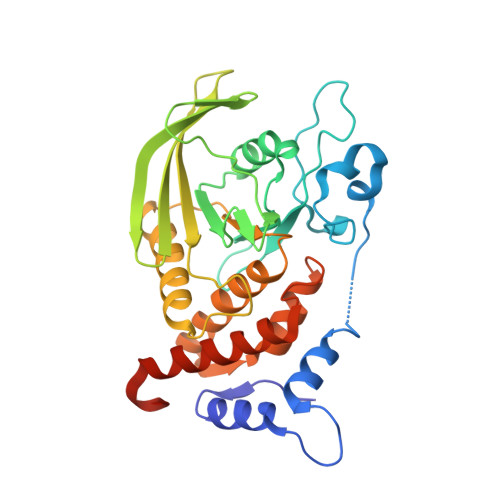
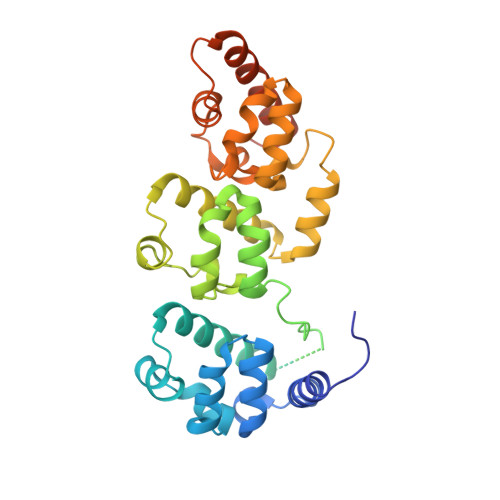
Structural insights into selective interaction between type IIa receptor protein tyrosine phosphatases and Liprin-alpha.
Wakita, M., Yamagata, A., Shiroshima, T., Izumi, H., Maeda, A., Sendo, M., Imai, A., Kubota, K., Goto-Ito, S., Sato, Y., Mori, H., Yoshida, T., Fukai, S.(2020) Nat Commun 11: 649-649
- PubMed: 32005855
- DOI: https://doi.org/10.1038/s41467-020-14516-5
- Primary Citation of Related Structures:
6KIP - PubMed Abstract:
Synapse formation is induced by transsynaptic interaction of neuronal cell-adhesion molecules termed synaptic organizers. Type IIa receptor protein tyrosine phosphatases (IIa RPTPs) function as presynaptic organizers. The cytoplasmic domain of IIa RPTPs consists of two phosphatase domains, and the membrane-distal one (D2) is essential for synapse formation. Liprin-α, which is an active zone protein critical for synapse formation, interacts with D2 via its C-terminal domain composed of three tandem sterile alpha motifs (tSAM). Structural mechanisms of this critical interaction for synapse formation remain elusive. Here, we report the crystal structure of the complex between mouse PTPδ D2 and Liprin-α3 tSAM at 1.91 Å resolution. PTPδ D2 interacts with the N-terminal helix and the first and second SAMs (SAM1 and SAM2, respectively) of Liprin-α3. Structure-based mutational analyses in vitro and in cellulo demonstrate that the interactions with Liprin-α SAM1 and SAM2 are essential for the binding and synaptogenic activity.
- Institute for Quantitative Biosciences, The University of Tokyo, Tokyo, 113-0032, Japan.
Organizational Affiliation: