Functional and crystallographic analysis of hydroxynitrile lyase from the millipede, Chamberlinius hualienensis
Motojima, F., Izumi, A., Zhai, Z., Yamaguchi, T., Shichida, S., Nakano, S., Dadashipour, M., Asano, Y.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hydroxynitrile lyase | 162 | Chamberlinius hualienensis | Mutation(s): 0 Gene Names: ChuaHNL | 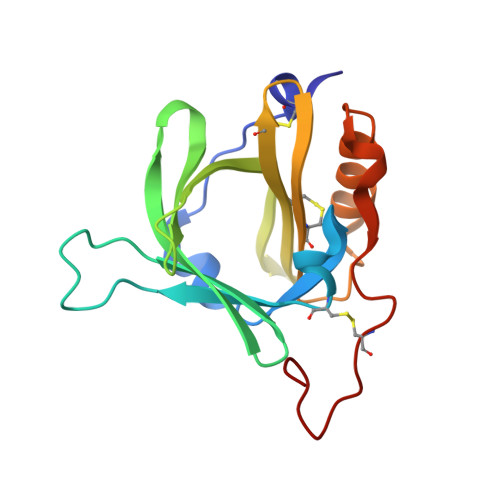 | |
UniProt | |||||
Find proteins for A0A0H5BR52 (Chamberlinius hualienensis) Explore A0A0H5BR52 Go to UniProtKB: A0A0H5BR52 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0H5BR52 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | D [auth A], E [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| ACT Query on ACT | B [auth A], C [auth A] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 57.924 | α = 90 |
| b = 57.924 | β = 90 |
| c = 226.321 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| SCALA | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Science and Technology | Japan | JPMJER1102 |