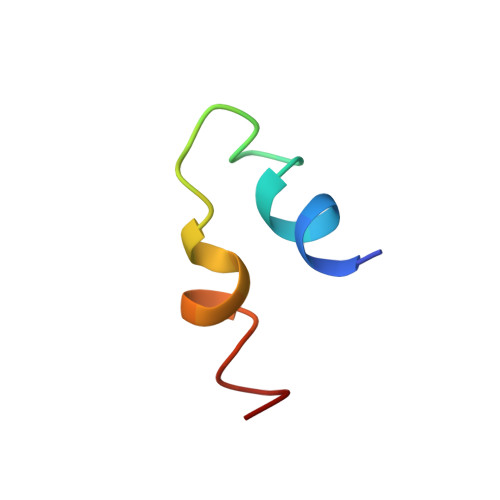
Structural insights into the combinatorial effects of antimicrobial peptides reveal a role of aromatic-aromatic interactions in antibacterial synergism.
Ilyas, H., Kim, J., Lee, D., Malmsten, M., Bhunia, A.(2019) J Biological Chem 294: 14615-14633
- PubMed: 31383740
- DOI: https://doi.org/10.1074/jbc.RA119.009955
- Primary Citation of Related Structures:
6KBO, 6KBV - PubMed Abstract:
The recent development of plants that overexpress antimicrobial peptides (AMPs) provides opportunities for controlling plant diseases. Because plants employ a broad-spectrum antimicrobial defense, including those based on AMPs, transgenic modification for AMP overexpression represents a potential way to utilize a defense system already present in plants. Herein, using an array of techniques and approaches, we report on VG16KRKP and KYE28, two antimicrobial peptides, which in combination exhibit synergistic antimicrobial effects against plant pathogens and are resistant against plant proteases. Investigating the structural origin of these synergistic antimicrobial effects with NMR spectroscopy of the complex formed between these two peptides and their mutated analogs, we demonstrate the formation of an unusual peptide complex, characterized by the formation of a bulky hydrophobic hub, stabilized by aromatic zippers. Using three-dimensional structure analyses of the complex in bacterial outer and inner membrane components and when bound to lipopolysaccharide (LPS) or bacterial membrane mimics, we found that this structure is key for elevating antimicrobial potency of the peptide combination. We conclude that the synergistic antimicrobial effects of VG16KRKP and KYE28 arise from the formation of a well-defined amphiphilic dimer in the presence of LPS and also in the cytoplasmic bacterial membrane environment. Together, these findings highlight a new application of solution NMR spectroscopy to solve complex structures to study peptide-peptide interactions, and they underscore the importance of structural insights for elucidating the antimicrobial effects of AMP mixtures.
- Department of Biophysics, Bose Institute, P-1/12 CIT Scheme VII (M), Kolkata 700054, India.
Organizational Affiliation: