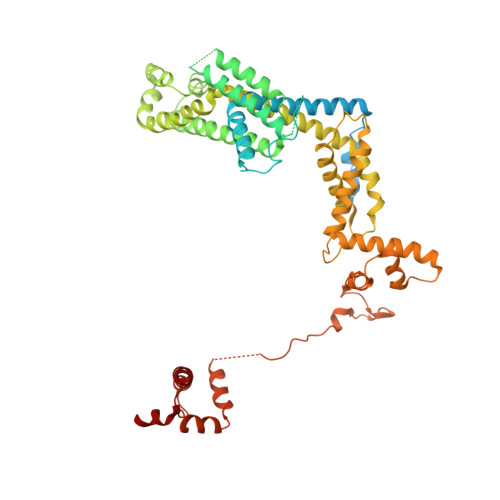
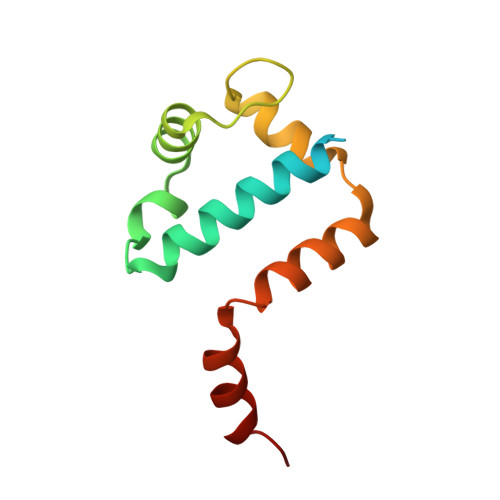
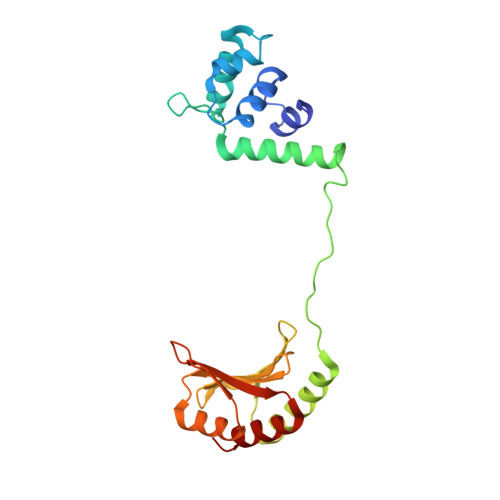
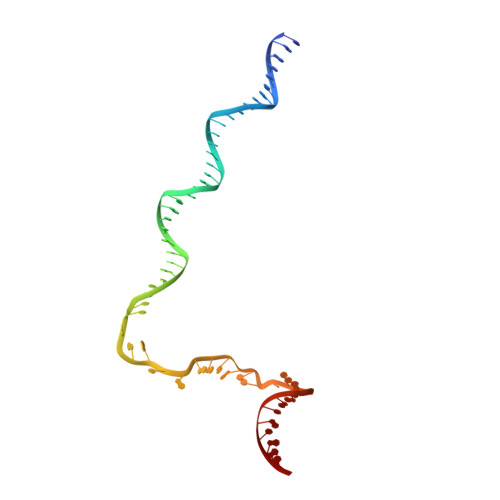
Structural basis of sigma appropriation.
Shi, J., Wen, A., Zhao, M., You, L., Zhang, Y., Feng, Y.(2019) Nucleic Acids Res 47: 9423-9432
- PubMed: 31392983
- DOI: https://doi.org/10.1093/nar/gkz682
- Primary Citation of Related Structures:
6K4Y - PubMed Abstract:
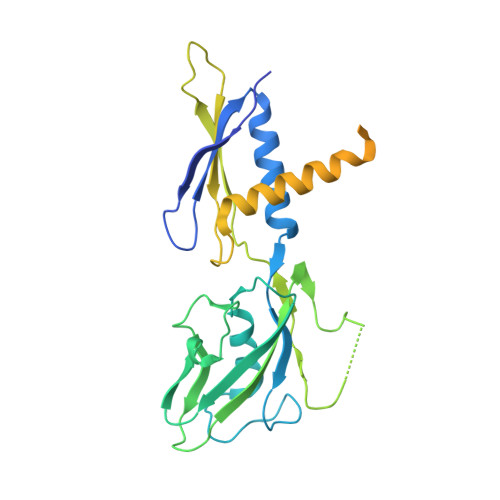
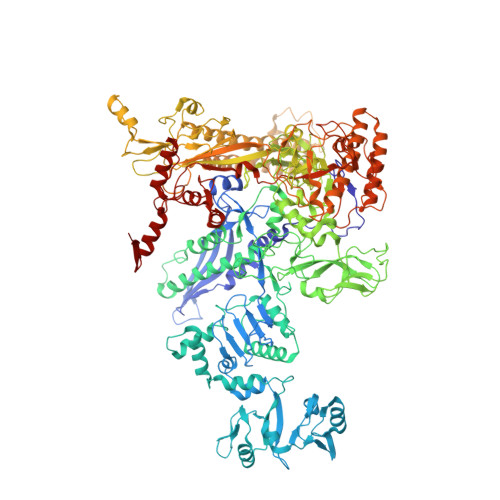
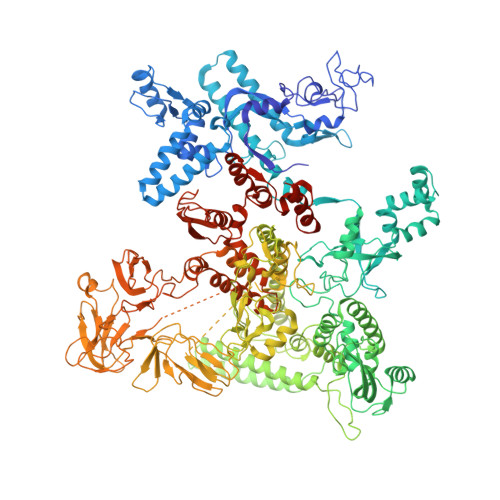
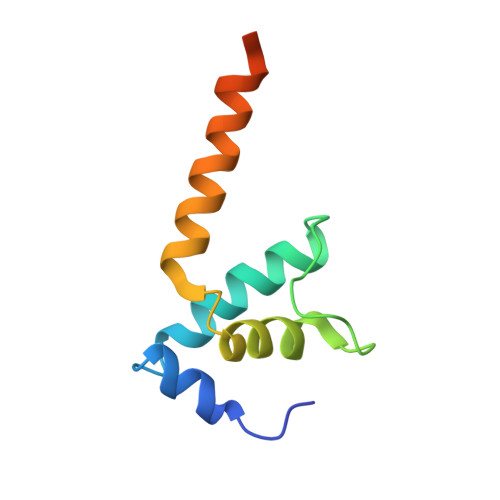
Bacteriophage T4 middle promoters are activated through a process called σ appropriation, which requires the concerted effort of two T4-encoded transcription factors: AsiA and MotA. Despite extensive biochemical and genetic analyses, puzzle remains, in part, because of a lack of precise structural information for σ appropriation complex. Here, we report a single-particle cryo-electron microscopy (cryo-EM) structure of an intact σ appropriation complex, comprising AsiA, MotA, Escherichia coli RNA polymerase (RNAP), σ70 and a T4 middle promoter. As expected, AsiA binds to and remodels σ region 4 to prevent its contact with host promoters. Unexpectedly, AsiA undergoes a large conformational change, takes over the job of σ region 4 and provides an anchor point for the upstream double-stranded DNA. Because σ region 4 is conserved among bacteria, other transcription factors may use the same strategy to alter the landscape of transcription immediately. Together, the structure provides a foundation for understanding σ appropriation and transcription activation.
- Department of Biophysics, and Department of Pathology of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou 310058, China.
Organizational Affiliation: