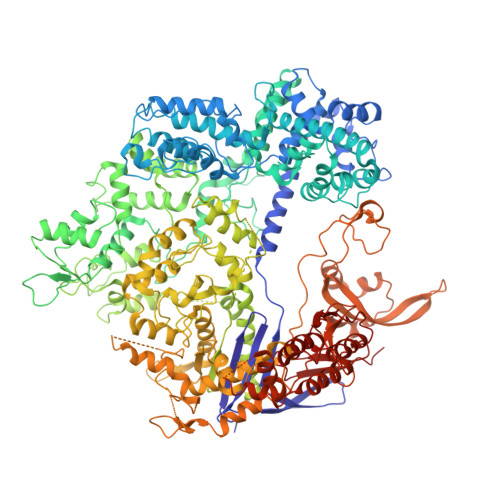
Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Chen, W., Zhang, H., Zhang, Y., Wang, Y., Gan, J., Ji, Q.(2019) PLoS Biol 17: e3000496-e3000496
- PubMed: 31603896
- DOI: https://doi.org/10.1371/journal.pbio.3000496
- Primary Citation of Related Structures:
6K3Z, 6K4P, 6K4Q, 6K4S, 6K4U, 6K57 - PubMed Abstract:
Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas systems have been harnessed as powerful genome editing tools in diverse organisms. However, the off-target effects and the protospacer adjacent motif (PAM) compatibility restrict the therapeutic applications of these systems. Recently, a Streptococcus pyogenes Cas9 (SpCas9) variant, xCas9, was evolved to possess both broad PAM compatibility and high DNA fidelity. Through determination of multiple xCas9 structures, which are all in complex with single-guide RNA (sgRNA) and double-stranded DNA containing different PAM sequences (TGG, CGG, TGA, and TGC), we decipher the molecular mechanisms of the PAM expansion and fidelity enhancement of xCas9. xCas9 follows a unique two-mode PAM recognition mechanism. For non-NGG PAM recognition, xCas9 triggers a notable structural rearrangement in the DNA recognition domains and a rotation in the key PAM-interacting residue R1335; such mechanism has not been observed in the wild-type (WT) SpCas9. For NGG PAM recognition, xCas9 applies a strategy similar to WT SpCas9. Moreover, biochemical and cell-based genome editing experiments pinpointed the critical roles of the E1219V mutation for PAM expansion and the R324L, S409I, and M694I mutations for fidelity enhancement. The molecular-level characterizations of the xCas9 nuclease provide critical insights into the mechanisms of the PAM expansion and fidelity enhancement of xCas9 and could further facilitate the engineering of SpCas9 and other Cas9 orthologs.
- School of Physical Science and Technology, ShanghaiTech University, Shanghai, China.
Organizational Affiliation: