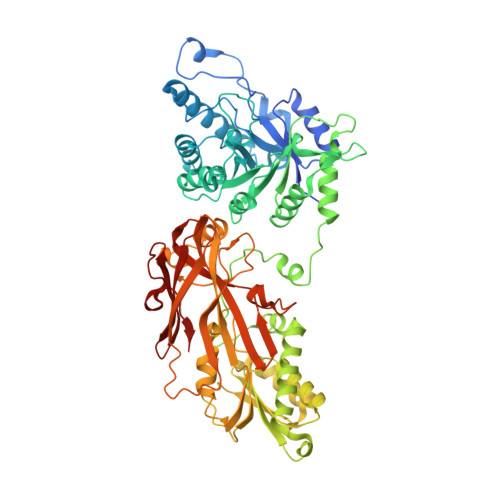
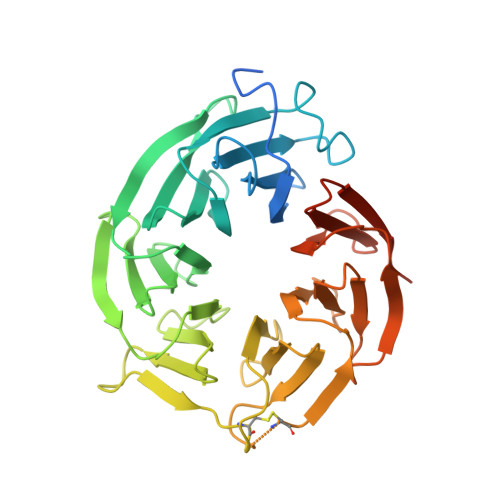
Discovery of Potent and Selective Covalent Protein Arginine Methyltransferase 5 (PRMT5) Inhibitors.
Lin, H., Wang, M., Zhang, Y.W., Tong, S., Leal, R.A., Shetty, R., Vaddi, K., Luengo, J.I.(2019) ACS Med Chem Lett 10: 1033-1038
- PubMed: 31312404
- DOI: https://doi.org/10.1021/acsmedchemlett.9b00074
- Primary Citation of Related Structures:
6K1S - PubMed Abstract:
Protein arginine methyltransferase 5 (PRMT5) is known to symmetrically dimethylate numerous cytosolic and nuclear proteins that are involved in a variety of cellular processes. Recent findings have revealed its potential as a cancer therapeutic target. PRMT5 possesses a cysteine (C449) in the active site, unique to PRMT5. Therefore, covalent PRMT5 inhibition is an attractive chemical approach. Herein, we report an exciting discovery of a series of novel hemiaminals that under physiological conditions can be converted to aldehydes and react with C449 to form covalent adducts, which presumably undergo an unprecedented elimination to form the thiol-vinyl ethers, as indicated by electron density in the co-crystal structure of the PRMT5/MEP50 complex.
- Prelude Therapeutics, 200 Powder Mill Road, Wilmington, Delaware 19803, United States.
Organizational Affiliation: