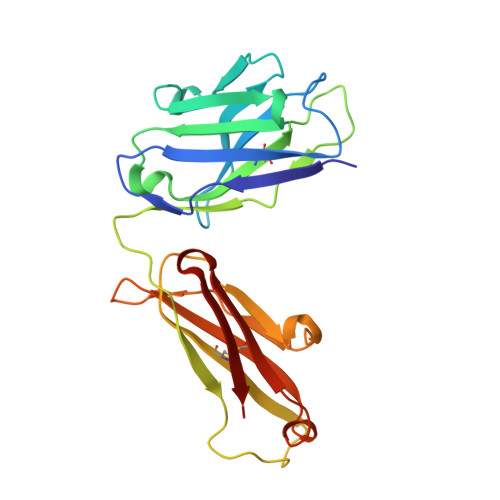
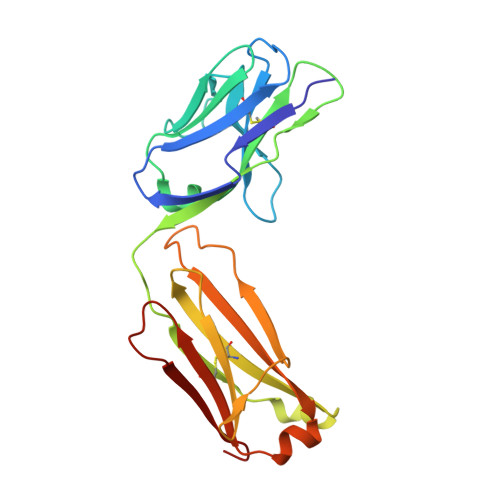
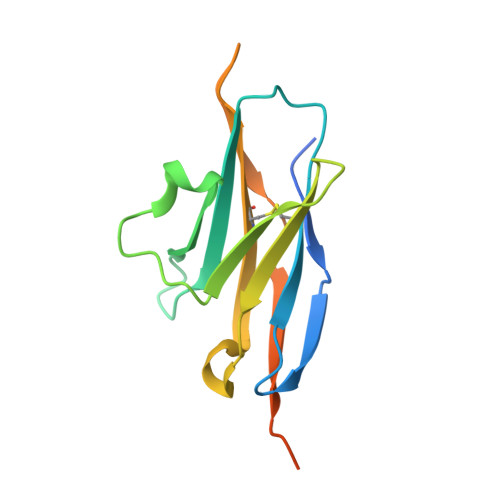
Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor.
Liu, J., Wang, G., Liu, L., Wu, R., Wu, Y., Fang, C., Zhou, X., Jiao, J., Gu, Y., Zhou, H., Xie, Z., Sun, Z., Chen, D., Dai, K., Wang, D., Tang, W., Yang, T.T.C.(2019) Sci Rep 9: 17830-17830
- PubMed: 31780710
- DOI: https://doi.org/10.1038/s41598-019-54231-w
- Primary Citation of Related Structures:
6K0Y - PubMed Abstract:
Programmed cell death 1 (PD-1) monoclonal antibodies have been approved by regulatory agencies for the treatment of various types of cancer, and the mechanism involves the restoration of T cell functions. We report herein the X-ray crystal structure of a fully human monoclonal antibody mAb059c fragment antigen-binding (Fab) in complex with the PD-1 extracellular domain (ECD) at a resolution of 1.70 Å. Structural analysis indicates 1) an epitope, comprising fragments from the C'D, BC and FG loops of PD-1, contributes to mAb059c interaction, 2) an unique conformation of the C'D loop and a different orientation of R86 enabling the capture of PD-1 by the antibody complementarity determining region (CDR) and the formation of one salt-bridge contact - ASP101(HCDR3):ARG86(PD-1), and 3) the contact of FG with light chain (LC) CDR3 is maintained by a second salt-bridge and two backbone hydrogen bonds. Interface analysis reveals that N-glycosylation sites 49, 74 and 116 on PD-1 do not contact mAb059c; while N58 in the BC loop is recognized by mAb059c heavy chain CDR1 and CDR2. Mutation of N58 attenuated mAb059c binding to PD-1. These findings and the novel anti-PD-1 antibody will facilitate better understanding of the mechanisms of the molecular recognition of PD-1 receptor by anti-PD-1 mAb and, thereby, enable the development of new therapeutics with an expanded spectrum of efficacy for unmet medical needs.
- Shanghai ChemPartner Co.Ltd, Shanghai, 201203, The People's Republic of China. jxliu@chempartner.com.
Organizational Affiliation: