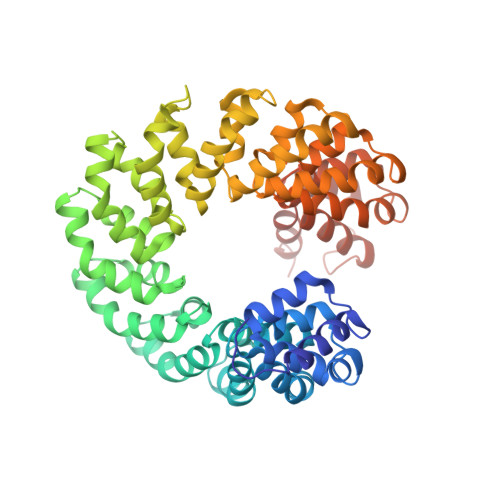
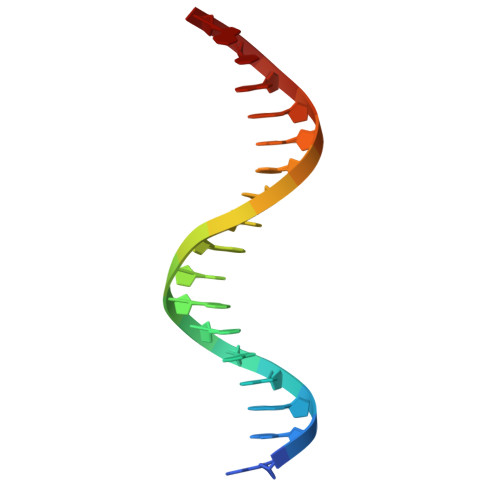
Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
Liu, L., Zhang, Y., Liu, M., Wei, W., Yi, C., Peng, J.(2020) J Mol Biology 432: 1035-1047
- PubMed: 31863750
- DOI: https://doi.org/10.1016/j.jmb.2019.11.023
- Primary Citation of Related Structures:
6JVZ, 6JW0, 6JW1, 6JW2, 6JW3, 6JW4, 6JW5 - PubMed Abstract:
Transcription activator-like effectors (TALEs) recognize DNA through repeat-variable diresidues (RVDs), and TALE-DNA interactions are sensitive to DNA modifications. Our previous study deciphered the recognition of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) by TALEs. Here, we report seven crystal structures of TALE-DNA complexes. The 5mC-specific RVD HA recognizes 5mC through van der Waals interactions and exhibits highly similar loop conformation to natural RVDs. The degenerate RVD RG contacts 5mC and 5hmC via van der Waals interactions as well; however, its loop conformation differs significantly. The loop conformations of universal RVD R* and 5hmC-specific RVD Q* are similar to that of RG, while the interactions of R* with C/5mC/5hmC and Q* with 5hmC are mediated by waters. Together, our findings illustrate the molecular basis for the specific recognition of 5mC and 5hmC by multiple noncanonical TALEs and provide insights into the plasticity of the TALE RVD loops.
- Academy for Advanced Interdisciplinary Studies, Peking University, Beijing 100871, China; Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China.
Organizational Affiliation: