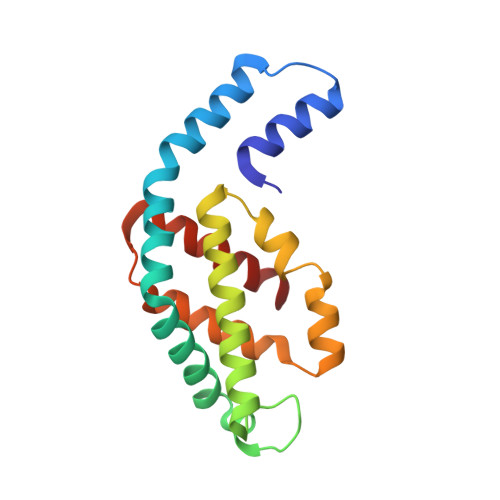
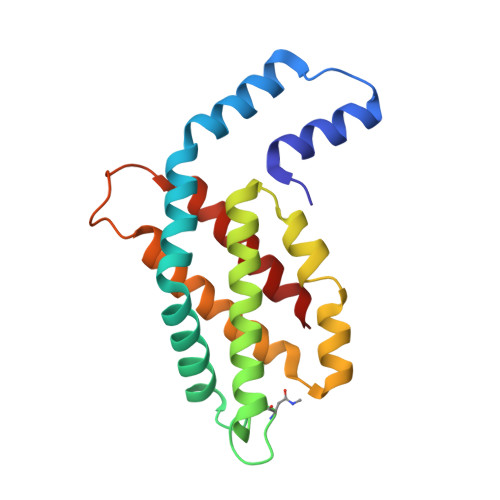
Phylogenetic and crystallographic analysis of Nostoc phycocyanin having blue-shifted spectral properties.
Sonani, R.R., Rastogi, R.P., Patel, S.N., Chaubey, M.G., Singh, N.K., Gupta, G.D., Kumar, V., Madamwar, D.(2019) Sci Rep 9: 9863-9863
- PubMed: 31285455
- DOI: https://doi.org/10.1038/s41598-019-46288-4
- Primary Citation of Related Structures:
6JPR - PubMed Abstract:
The distinct sequence feature and spectral blue-shift (~10 nm) of phycocyanin, isolated from Nostoc sp. R76DM (N-PC), were investigated by phylogenetic and crystallographic analyses. Twelve conserved substitutions in N-PC sequence were found distributed unequally among α- and β-subunit (3 in α- and 9 in β-subunit). The phylogenetic analysis suggested that molecular evolution of α- and β-subunit of Nostoc-phycocyanin is faster than evolution of Nostoc-species. The divergence events seem to have occurred more frequently in β-subunit, compared to α-subunit (relative divergence, 7.38 for α-subunit and 9.66 for β-subunit). Crystal structure of N-PC was solved at 2.35 Å resolution to reasonable R-factors (R work /R Free = 0.199/0.248). Substitutions congregate near interface of two αβ-monomer in N-PC trimer and are of compensatory nature. Six of the substitutions in β-subunit may be involved in maintaining topology of β-subunit, one in inter-monomer interaction and one in interaction with linker-protein. The β153Cys-attached chromophore adopts high-energy conformational state resulting due to reduced coplanarity of B- and C-pyrrole rings. Distortion in chromophore conformation can result in blue-shift in N-PC spectral properties. N-PC showed significant in-vitro and in-vivo antioxidant activity comparable with other phycocyanin. Since Nostoc-species constitute a distinct phylogenetic clade, the present structure would provide a better template to build a model for phycocyanins of these species.
- Radiation Biology & Health Sciences Division, Bhabha Atomic Research Centre, Trombay, Mumbai, 400 085, India.
Organizational Affiliation: