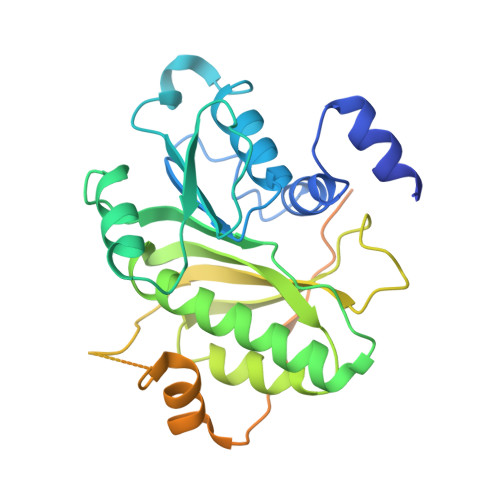
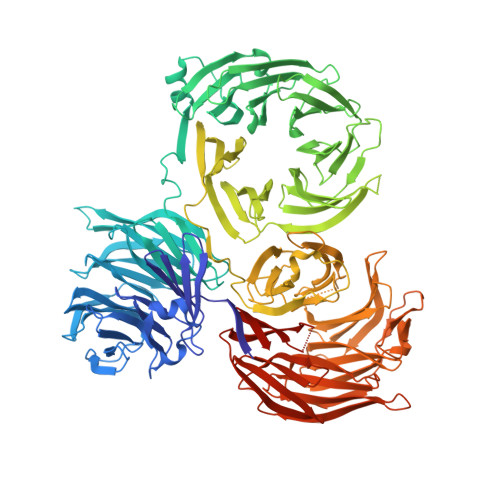Structure of tRNA methyltransferase complex of Trm7 and Trm734 reveals a novel binding interface for tRNA recognition.
Hirata, A., Okada, K., Yoshii, K., Shiraishi, H., Saijo, S., Yonezawa, K., Shimizu, N., Hori, H.(2019) Nucleic Acids Res 47: 10942-10955
- PubMed: 31586407
- DOI: https://doi.org/10.1093/nar/gkz856
- Primary Citation of Related Structures:
6JP6, 6JPL - PubMed Abstract:
The complex between Trm7 and Trm734 (Trm7-Trm734) from Saccharomyces cerevisiae catalyzes 2'-O-methylation at position 34 in tRNA. We report biochemical and structural studies of the Trm7-Trm734 complex. Purified recombinant Trm7-Trm734 preferentially methylates tRNAPhe transcript variants possessing two of three factors (Cm32, m1G37 and pyrimidine34). Therefore, tRNAPhe, tRNATrp and tRNALeu are specifically methylated by Trm7-Trm734. We have solved the crystal structures of the apo and S-adenosyl-L-methionine bound forms of Trm7-Trm734. Small angle X-ray scattering reveals that Trm7-Trm734 exists as a hetero-dimer in solution. Trm7 possesses a Rossmann-fold catalytic domain, while Trm734 consists of three WD40 β-propeller domains (termed BPA, BPB and BPC). BPA and BPC form a unique V-shaped cleft, which docks to Trm7. The C-terminal region of Trm7 is required for binding to Trm734. The D-arm of substrate tRNA is required for methylation by Trm7-Trm734. If the D-arm in tRNAPhe is docked onto the positively charged area of BPB in Trm734, the anticodon-loop is located near the catalytic pocket of Trm7. This model suggests that Trm734 is required for correct positioning of tRNA for methylation. Additionally, a point-mutation in Trm7, which is observed in FTSJ1 (human Trm7 ortholog) of nosyndromic X-linked intellectual disability patients, decreases the methylation activity.
- Department of Materials Science and Biotechnology, Graduate school of Science and Engineering, Ehime University, 3 Bunkyo-cho, Matsuyama, Ehime 790-8577, Japan.
Organizational Affiliation: