Structural basis for the promiscuous PAM recognition by Corynebacterium diphtheriae Cas9.
Hirano, S., Abudayyeh, O.O., Gootenberg, J.S., Horii, T., Ishitani, R., Hatada, I., Zhang, F., Nishimasu, H., Nureki, O.(2019) Nat Commun 10: 1968-1968
- PubMed: 31036811
- DOI: https://doi.org/10.1038/s41467-019-09741-6
- Primary Citation of Related Structures:
6JOO - PubMed Abstract:
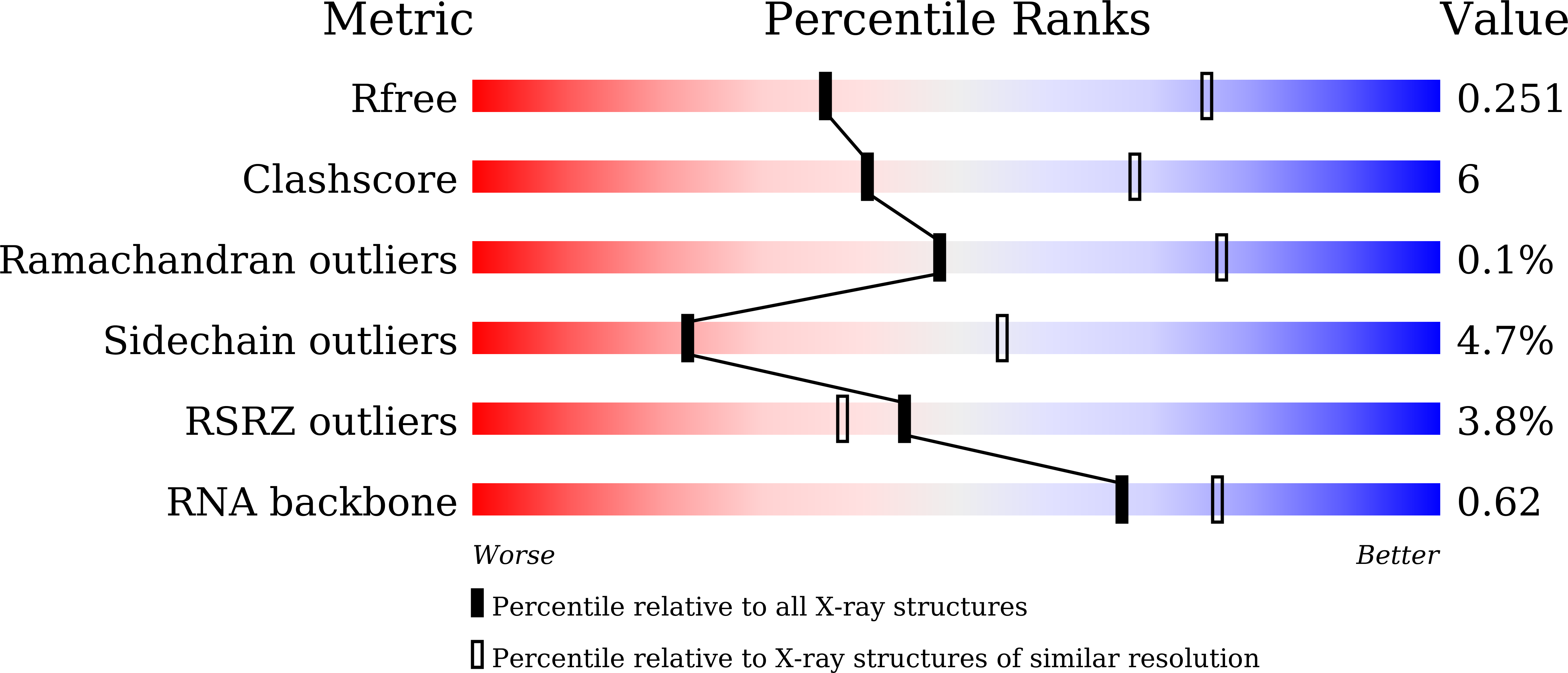
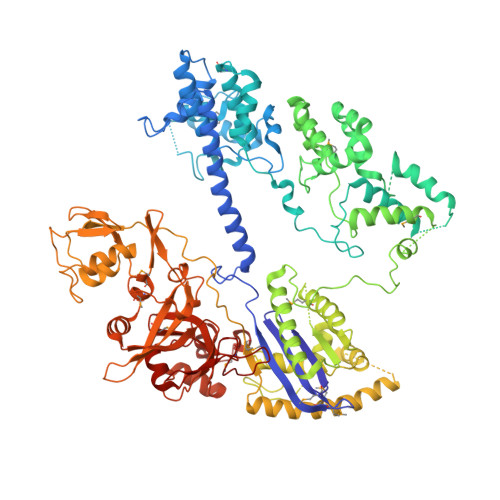
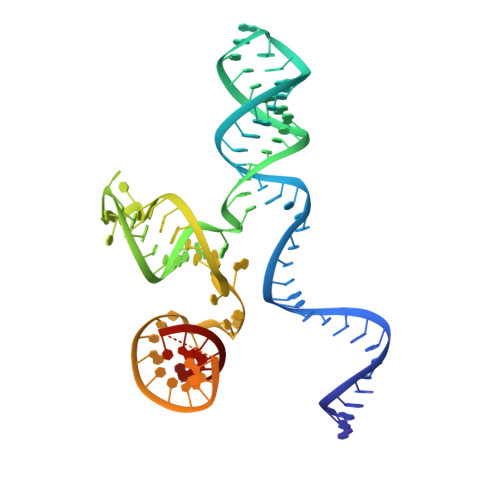
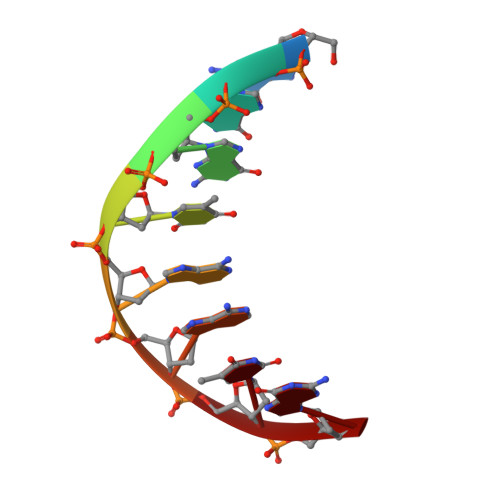
The RNA-guided DNA endonuclease Cas9 cleaves double-stranded DNA targets bearing a protospacer adjacent motif (PAM) and complementarity to an RNA guide. Unlike other Cas9 orthologs, Corynebacterium diphtheriae Cas9 (CdCas9) recognizes the promiscuous NNRHHHY PAM. However, the CdCas9-mediated PAM recognition mechanism remains unknown. Here, we report the crystal structure of CdCas9 in complex with the guide RNA and its target DNA at 2.9 Å resolution. The structure reveals that CdCas9 recognizes the NNRHHHY PAM via a combination of van der Waals interactions and base-specific hydrogen bonds. Moreover, we find that CdCas9 exhibits robust DNA cleavage activity with the optimal 22-nucleotide length guide RNAs. Our findings highlight the mechanistic diversity of the PAM recognition by Cas9 orthologs, and provide a basis for the further engineering of the CRISPR-Cas9 genome-editor nucleases.
- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo, 113-0033, Japan.
Organizational Affiliation: