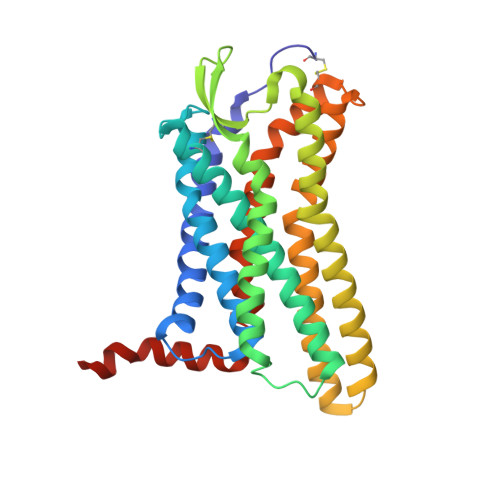
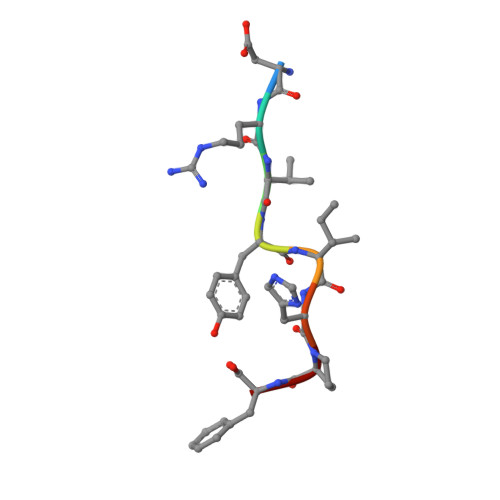
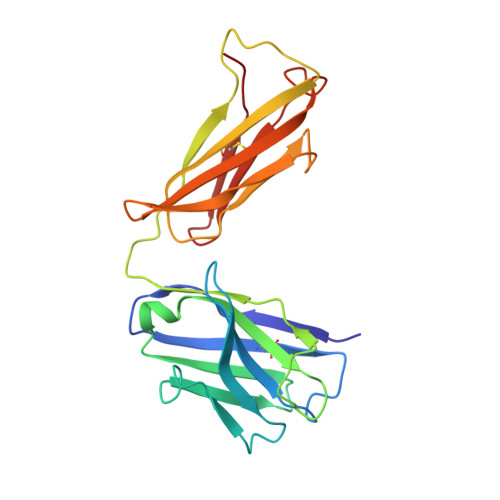
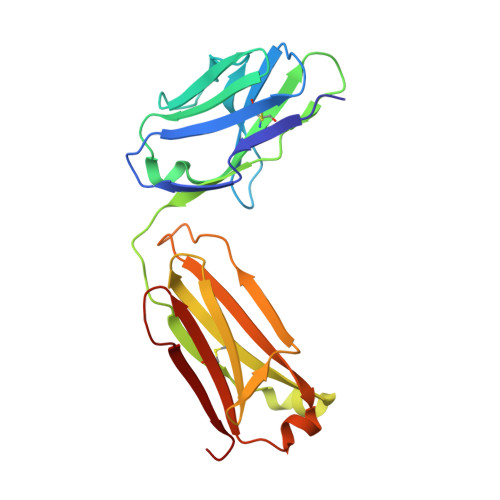
The Crystal Structure of Angiotensin II Type 2 Receptor with Endogenous Peptide Hormone.
Asada, H., Inoue, A., Ngako Kadji, F.M., Hirata, K., Shiimura, Y., Im, D., Shimamura, T., Nomura, N., Iwanari, H., Hamakubo, T., Kusano-Arai, O., Hisano, H., Uemura, T., Suno, C., Aoki, J., Iwata, S.(2020) Structure 28: 418
- PubMed: 31899086
- DOI: https://doi.org/10.1016/j.str.2019.12.003
- Primary Citation of Related Structures:
6JOD - PubMed Abstract:
Angiotensin II (AngII) is a peptide hormone that plays a key role in regulating blood pressure, and its interactions with the G protein-coupled receptors, AngII type-1 receptor (AT 1 R) and AngII type-2 receptor (AT 2 R), are central to its mechanism of action. We solved the crystal structure of human AT 2 R bound to AngII and its specific antibody at 3.2-Å resolution. AngII (full agonist) and [Sar 1 , Ile 8 ]-AngII (partial agonist) interact with AT 2 R in a similar fashion, except at the bottom of the AT 2 R ligand-binding pocket. In particular, the residues including Met128 3.36 , which constitute the deep end of the cavity, play important roles in angiotensin receptor (ATR) activation upon AngII binding. These differences that occur upon endogenous ligand binding may contribute to a structural change in AT 2 R, leading to normalization of the non-canonical coordination of helix 8. Our results will inform the design of more effective ligands for ATRs.
- Department of Cell Biology, Graduate School of Medicine, Kyoto University, Kyoto 606-8501, Japan. Electronic address: asada.hidetsugu.4s@kyoto-u.ac.jp.
Organizational Affiliation: