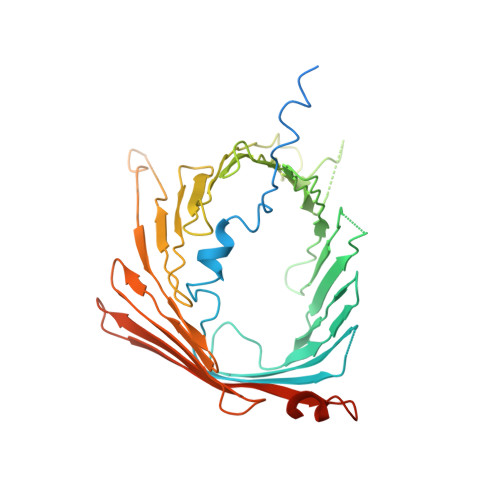
Structure of the mitochondrial import gate reveals distinct preprotein paths.
Araiso, Y., Tsutsumi, A., Qiu, J., Imai, K., Shiota, T., Song, J., Lindau, C., Wenz, L.S., Sakaue, H., Yunoki, K., Kawano, S., Suzuki, J., Wischnewski, M., Schutze, C., Ariyama, H., Ando, T., Becker, T., Lithgow, T., Wiedemann, N., Pfanner, N., Kikkawa, M., Endo, T.(2019) Nature 575: 395-401
- PubMed: 31600774
- DOI: https://doi.org/10.1038/s41586-019-1680-7
- Primary Citation of Related Structures:
6JNF - PubMed Abstract:
The translocase of the outer mitochondrial membrane (TOM) is the main entry gate for proteins 1-4 . Here we use cryo-electron microscopy to report the structure of the yeast TOM core complex 5-9 at 3.8-Å resolution. The structure reveals the high-resolution architecture of the translocator consisting of two Tom40 β-barrel channels and α-helical transmembrane subunits, providing insight into critical features that are conserved in all eukaryotes 1-3 . Each Tom40 β-barrel is surrounded by small TOM subunits, and tethered by two Tom22 subunits and one phospholipid. The N-terminal extension of Tom40 forms a helix inside the channel; mutational analysis reveals its dual role in early and late steps in the biogenesis of intermembrane-space proteins in cooperation with Tom5. Each Tom40 channel possesses two precursor exit sites. Tom22, Tom40 and Tom7 guide presequence-containing preproteins to the exit in the middle of the dimer, whereas Tom5 and the Tom40 N extension guide preproteins lacking a presequence to the exit at the periphery of the dimer.
- Faculty of Life Sciences, Kyoto Sangyo University, Kyoto, Japan.
Organizational Affiliation: