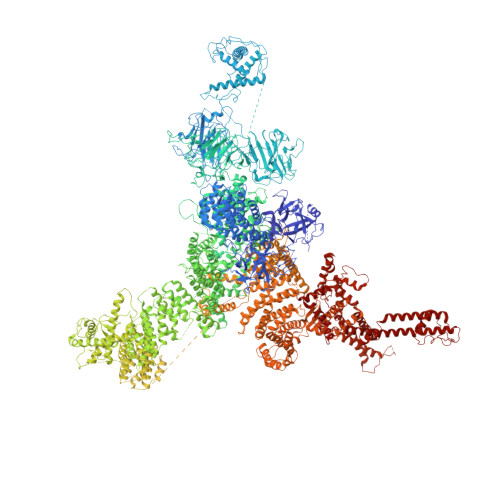
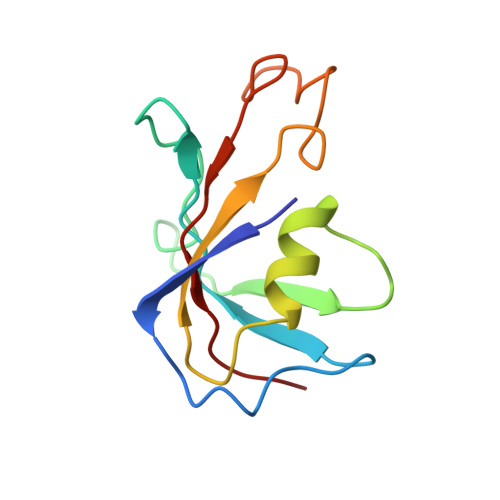
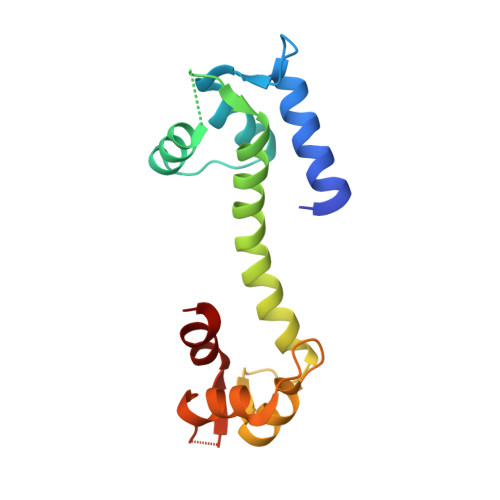
Modulation of cardiac ryanodine receptor 2 by calmodulin.
Gong, D., Chi, X., Wei, J., Zhou, G., Huang, G., Zhang, L., Wang, R., Lei, J., Chen, S.R.W., Yan, N.(2019) Nature 572: 347-351
- PubMed: 31278385
- DOI: https://doi.org/10.1038/s41586-019-1377-y
- Primary Citation of Related Structures:
6JI0, 6JI8, 6JII, 6JIU, 6JIY, 6JRR, 6JRS, 6JV2 - PubMed Abstract:
The high-conductance intracellular calcium (Ca 2+ ) channel RyR2 is essential for the coupling of excitation and contraction in cardiac muscle. Among various modulators, calmodulin (CaM) regulates RyR2 in a Ca 2+ -dependent manner. Here we reveal the regulatory mechanism by which porcine RyR2 is modulated by human CaM through the structural determination of RyR2 under eight conditions. Apo-CaM and Ca 2+ -CaM bind to distinct but overlapping sites in an elongated cleft formed by the handle, helical and central domains. The shift in CaM-binding sites on RyR2 is controlled by Ca 2+ binding to CaM, rather than to RyR2. Ca 2+ -CaM induces rotations and intradomain shifts of individual central domains, resulting in pore closure of the PCB95 and Ca 2+ -activated channel. By contrast, the pore of the ATP, caffeine and Ca 2+ -activated channel remains open in the presence of Ca 2+ -CaM, which suggests that Ca 2+ -CaM is one of the many competing modulators of RyR2 gating.
- Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing, China. gds13@tsinghua.org.cn.
Organizational Affiliation: