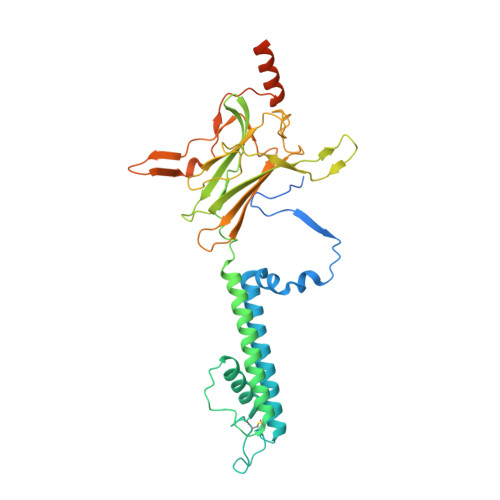
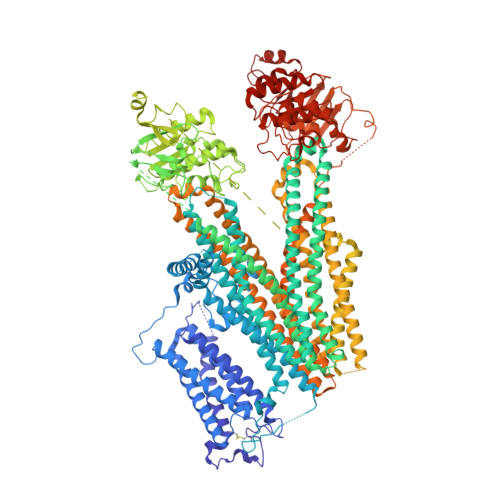
The Structural Basis for the Binding of Repaglinide to the Pancreatic KATPChannel.
Ding, D., Wang, M., Wu, J.X., Kang, Y., Chen, L.(2019) Cell Rep 27: 1848-1857.e4
- PubMed: 31067468
- DOI: https://doi.org/10.1016/j.celrep.2019.04.050
- Primary Citation of Related Structures:
6JB1, 6JB3 - PubMed Abstract:
Repaglinide (RPG) is a short-acting insulin secretagogue widely prescribed for the treatment of type 2 diabetes. It boosts insulin secretion by inhibiting the pancreatic ATP-sensitive potassium channel (K ATP ). However, the mechanisms by which RPG binds to the K ATP channel are poorly understood. Here, we describe two cryo-EM structures: the pancreatic K ATP channel in complex with inhibitory RPG and adenosine-5'-(γ-thio)-triphosphate (ATPγS) at 3.3 Å and a medium-resolution structure of a RPG-bound mini SUR1 protein in which the N terminus of the inward-rectifying potassium channel 6.1 (Kir6.1) is fused to the ABC transporter module of the sulfonylurea receptor 1 (SUR1). These structures reveal the binding site of RPG in the SUR1 subunit. Furthermore, the high-resolution structure reveals the complex architecture of the ATP binding site, which is formed by both Kir6.2 and SUR1 subunits, and the domain-domain interaction interfaces.
- State Key Laboratory of Membrane Biology, Institute of Molecular Medicine, Peking University, Beijing Key Laboratory of Cardiometabolic Molecular Medicine, Beijing 100871, China; Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China; Academy for Advanced Interdisciplinary Studies, Peking University, Beijing 100871, China.
Organizational Affiliation: