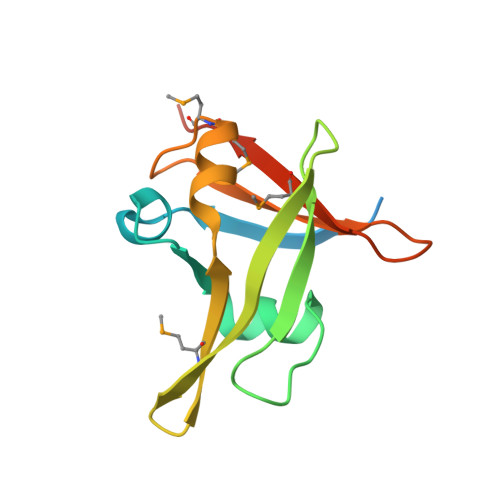
Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Tao, Z., Hu, H., Luo, X., Jia, B., Du, J., He, Y.(2019) Nat Plants 5: 424-435
- PubMed: 30962525
- DOI: https://doi.org/10.1038/s41477-019-0402-3
- Primary Citation of Related Structures:
6J9A, 6J9B, 6J9C - PubMed Abstract:
Some overwintering plants acquire competence to flower, after experiencing prolonged cold in winter, through a process termed vernalization. In the crucifer plant Arabidopsis thaliana, prolonged cold induces chromatin-mediated silencing of the potent floral repressor FLOWERING LOCUS C (FLC) by Polycomb proteins. This vernalized state is epigenetically maintained or 'memorized' in warm rendering plants competent to flower in spring, but is reset in the next generation. Here, we show that in early embryogenesis, two homologous B3 domain transcription factors LEAFY COTYLEDON 2 (LEC2) and FUSCA3 (FUS3) compete against two repressive B3-containing epigenome readers and Polycomb partners known as VAL1 and VAL2 for the cis-regulatory cold memory element (CME) of FLC to disrupt Polycomb silencing. Consistently, crystal structures of B3-CME complexes show that B3 FUS3 , B3 LEC2 and B3 VAL1 employ a nearly identical binding interface for CME. We further found that LEC2 and FUS3 recruit the scaffold protein FRIGIDA in association with active chromatin modifiers to establish an active chromatin state at FLC, which results in resetting of the silenced FLC to active and erasing the epigenetic parental memory of winter cold in early embryos. Following embryo development, LEC2 and FUS3 are developmentally silenced throughout post-embryonic stages, enabling VALs to bind to the CME again at seedling stages at which plants experience winter cold. Our findings illustrate how overwintering crucifer annuals or biennials in temperate climates employ a subfamily of B3 domain proteins to switch on, off and on again the expression of a key flowering gene in the embryo-to-plant-to-embryo cycle, and thus to synchronize growth and development with seasonal temperature changes in their life cycles.
- Shanghai Center for Plant Stress Biology & National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences (CAS), Shanghai, China.
Organizational Affiliation: