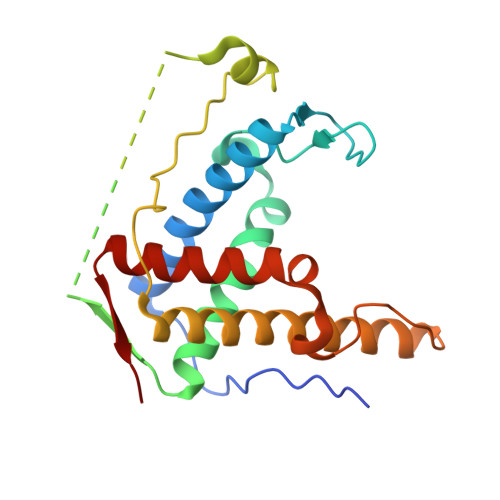
Crystal structure of diamondback moth ryanodine receptor Repeat34 domain reveals insect-specific phosphorylation sites.
Xu, T., Yuchi, Z.(2019) BMC Biol 17: 77-77
- PubMed: 31597572
- DOI: https://doi.org/10.1186/s12915-019-0698-5
- Primary Citation of Related Structures:
6J6O, 6J6P - PubMed Abstract:
Ryanodine receptor (RyR), a calcium-release channel located in the sarcoplasmic reticulum membrane of muscles, is the target of insecticides used against a wide range of agricultural pests. Mammalian RyRs have been shown to be under the regulatory control of several kinases and phosphatases, but little is known about the regulation of insect RyRs by phosphorylation. Here we present the crystal structures of wild-type and phospho-mimetic RyR Repeat34 domain containing PKA phosphorylation sites from diamondback moth (DBM), a major lepidopteran pest of cruciferous vegetables. The structure has unique features, not seen in mammalian RyRs, including an additional α-helix near the phosphorylation loop. Using tandem mass spectrometry, we identify several PKA sites clustering in the phosphorylation loop and the newly identified α-helix. Bioinformatics analysis shows that this α-helix is only present in Lepidoptera, suggesting an insect-specific regulation. Interestingly, the specific phosphorylation pattern is temperature-dependent. The thermal stability of the DBM Repeat34 domain is significantly lower than that of the analogous domain in the three mammalian RyR isoforms, indicating a more dynamic domain structure that can be partially unfolded to facilitate the temperature-dependent phosphorylation. Docking the structure into the cryo-electron microscopy model of full-length RyR reveals that the interface between the Repeat34 and neighboring HD1 domain is more conserved than that of the phosphorylation loop region that might be involved in the interaction with SPRY3 domain. We also identify an insect-specific glycerol-binding pocket that could be potentially targeted by novel insecticides to fight the current resistance crisis. The crystal structures of the DBM Repeat34 domain reveals insect-specific temperature-dependent phosphorylation sites that may regulate insect ryanodine receptor function. It also reveals insect-specific structural features and a potential ligand-binding site that could be targeted in an effort to develop green pesticides with high species-specificity.
- Tianjin Key Laboratory for Modern Drug Delivery & High-Efficiency, Collaborative Innovation Center of Chemical Science and Engineering, School of Pharmaceutical Science and Technology, Tianjin University, Tianjin, 300072, China.
Organizational Affiliation: